Transcription factor binding site prediction method fusing with DNA shape features
A shape feature and binding site technology, applied in the field of bioinformatics, can solve problems such as good prediction of TF binding preference, and achieve the effects of strong usability, low design complexity, and short training time
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
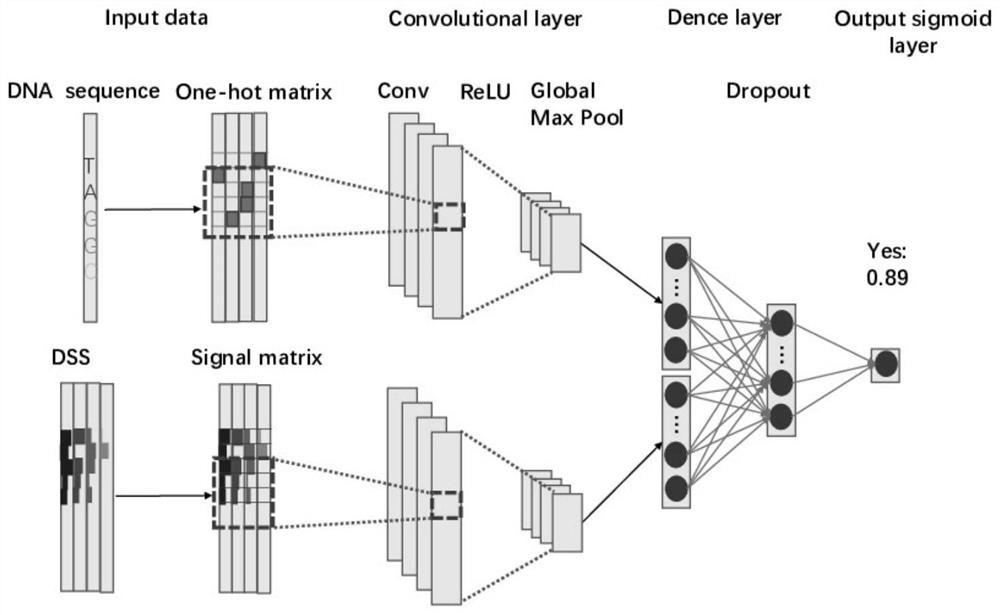
[0028] Step A fusion DNA specific shape features transcription factor binding site prediction, the method bits are as follows:
[0029] Construction 1, the data set
[0030] First, in-depth study of the underlying mechanisms related to scientific progress -DNA binding proteins, as well as summarize the current status and recent development of DNA transcription factor binding site prediction, collect survey data sets mainstream sources of information in the field. Secondly, the fusion DNA shape characteristics and sequence information transcription factor binding site prediction model, in-depth study DNA shape characteristics acquired relevant developments methods, as well as DNA transcription factor binding site prediction related data set construction method, designed and constructed with DNA shape specific characteristic data and the data set of DNA sequence information.
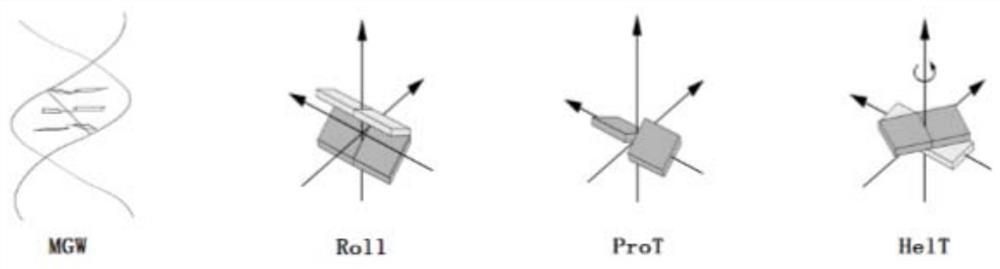
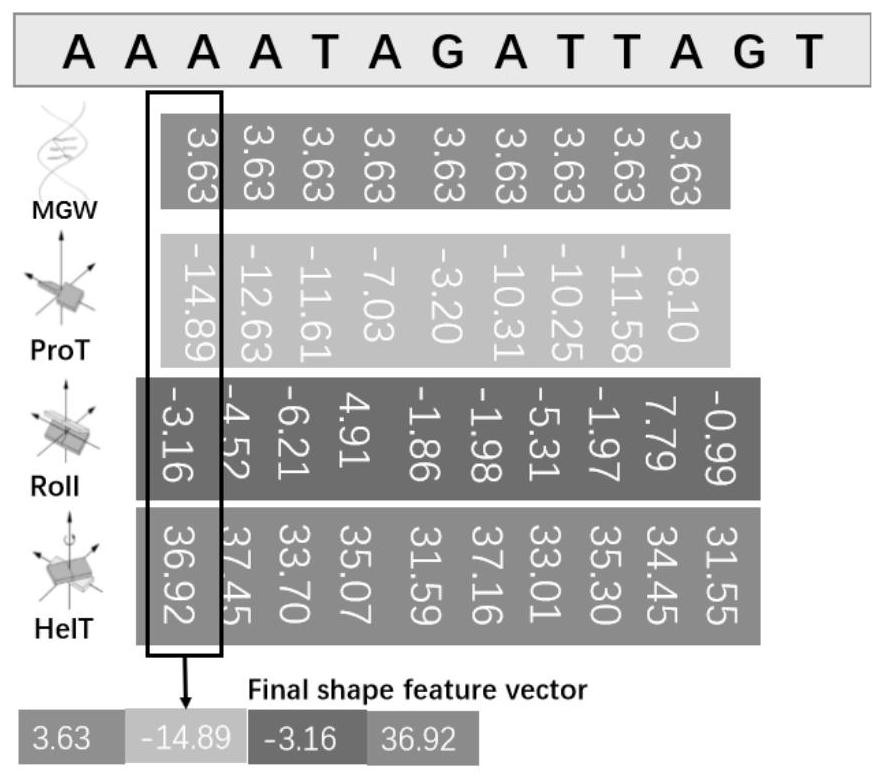
[0031]Method using HT-MC DNA shape characteristics acquired, previous studies to improve efficiency by redu...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com