DNA metabarcoding detection target sequence, detection kit and detection method for screening animal provenance components in meat products
A detection kit and macro-barcode technology, which is applied in the field of DNA macro-barcode detection target sequence, can solve the problems of difficult design, poor specificity, and low sensitivity, so as to avoid false negative results, solve the difficulty of blind detection, and improve reliability Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] 1.1. DNA metabarcode detection target sequence screening and universal primer design
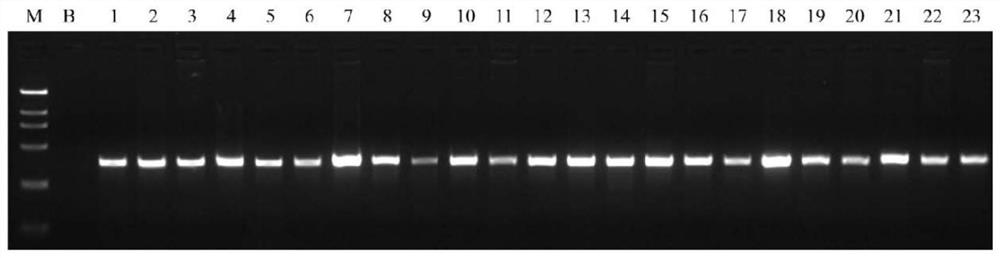
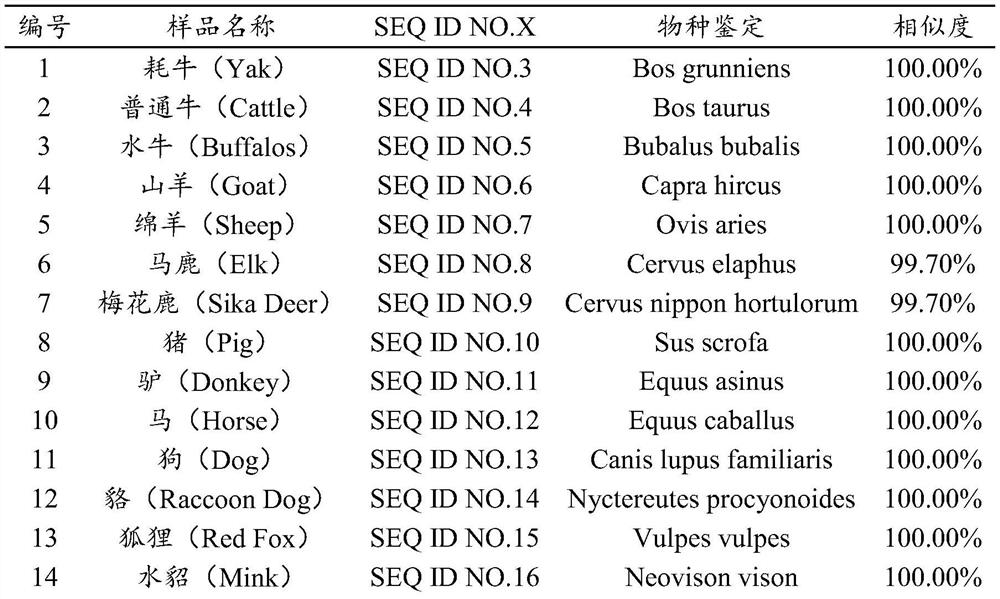
[0044] Download 23 target species from NCBI's Nucleotide database (yak, common cattle, buffalo, goat, sheep, red deer, sika deer, pig, donkey, horse, dog, raccoon dog, fox, mink, cat, rabbit, guinea pig, mouse, Rat, hamster, chicken, duck, goose) target sequence, and use MEGA software for sequence alignment analysis, and design a pair of universal primers in the conserved region. The forward primer region contains three variant sites in the reference genomes of 23 species. The design uses degenerate bases when synthesizing the variant sites. The base M is synthesized according to the degenerate base A / C, and the base Y is synthesized according to the degenerate base A / C. The degenerate base C / T is synthesized, the base R is synthesized according to the degenerate base A / G, and the reverse primer region is completely conserved, and the amplified region has a high degree of interspecies...
Embodiment 2
[0062] Detection of target species-derived components in the sample of Example 2
[0063] 2.1. Sample collection and DNA extraction
[0064] Sample collection and DNA extraction were the same as in Example 1.
[0065] 2.2. Sample preparation
[0066] The DNA of 23 species was mixed in the same proportion, each species’ DNA content was 4.35%, and the final concentration was about 0.87 ng / μL. It was used as a test sample and stored at -20°C for future use.
[0067] 2.3.PCR amplification and product recovery and purification
[0068] PCR amplification is the same as in Example 1.
[0069] Purification of PCR products using Gel DNA Extraction Mini Kit kit or other recognized methods for purification of amplified products with the same efficacy.
[0070] 2.4. Next Generation Sequencing
[0071] Illumina library construction kit was used to construct the sequencing library of the purified amplified products, and the Illumina NovaSeq PE250 platform was used for next-generation...
Embodiment 3
[0084] Embodiment 3 sensitivity test
[0085] The DNA of 23 species is mixed in different proportions, and 14 food animals (yak, common cattle, buffalo, goat, sheep, red deer, sika deer, pig, donkey, horse, rabbit, chicken, duck, goose) are used as the main ingredients , the proportion of DNA in each species was 7.08%, and the final concentration was about 1.42ng / μL; 9 non-edible animals (dog, raccoon dog, fox, mink, cat, guinea pig, mouse, rat, hamster) were used as secondary Components, the proportion of DNA of each species is 0.10%, the final concentration is about 0.02ng / μL.
[0086] The PCR amplification is the same as in Example 1, and the recovery and purification of PCR amplification products, next-generation sequencing, and bioinformatics analysis are the same as in Example 2. The test results are shown in Table 3. The test results showed that after the sequencing data were analyzed by bioinformatics, all species components in the DNA mixed samples were identified. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com