Rapid screening method for hepatotoxic compounds in polygonum multiflorum based on MRP2/3
A compound and hepatotoxicity technology, which is applied in the field of rapid screening of hepatotoxic compounds in Polygonum multiflorum, can solve problems affecting chemotherapy drug resistance and other issues
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] Example 1 Computer Molecular Docking Screening of Hepatotoxic Compounds
[0061] The main components of Radix Polygoni Multiflori were collected through literature and database retrieval, and a total of 49 molecules were obtained. DiscoveryStudio2.5 was used to preprocess these molecules. The preprocessing mainly included the following operations: three-dimensional structure generation, structure optimization, and repeated structure deletion. 49 molecules were obtained. Chemical structures, based on which ligand libraries are constructed and subsequently used for virtual screening.
[0062] The Swiss-model database was searched with the keywords "MRP2" and "MRP3", and a homology model structure was obtained according to the screening rules. The template PDB number of MRP2 and MRP3 was 6uy0. According to literature [1-2] The recorded amino acid residues define the active site, and MRP2 has a radius of 位点的三维坐标位置为157.158707,176.372707,163.243586,其关键氨基酸残基为LYS295、VAL434、S...
Embodiment 2
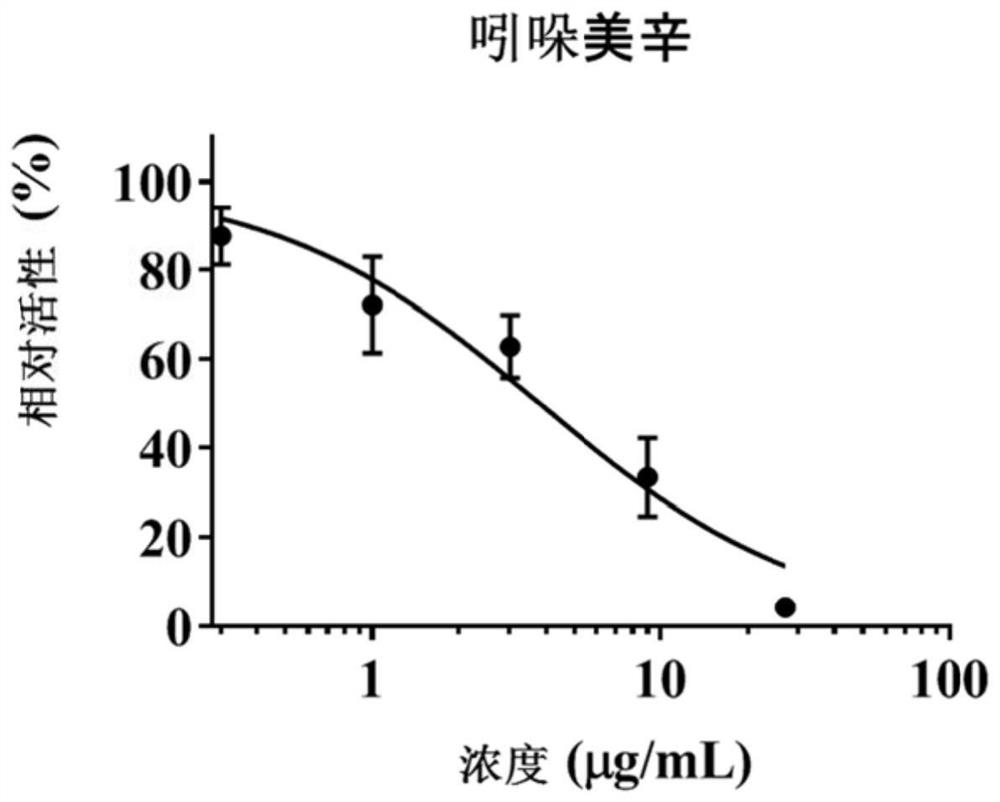
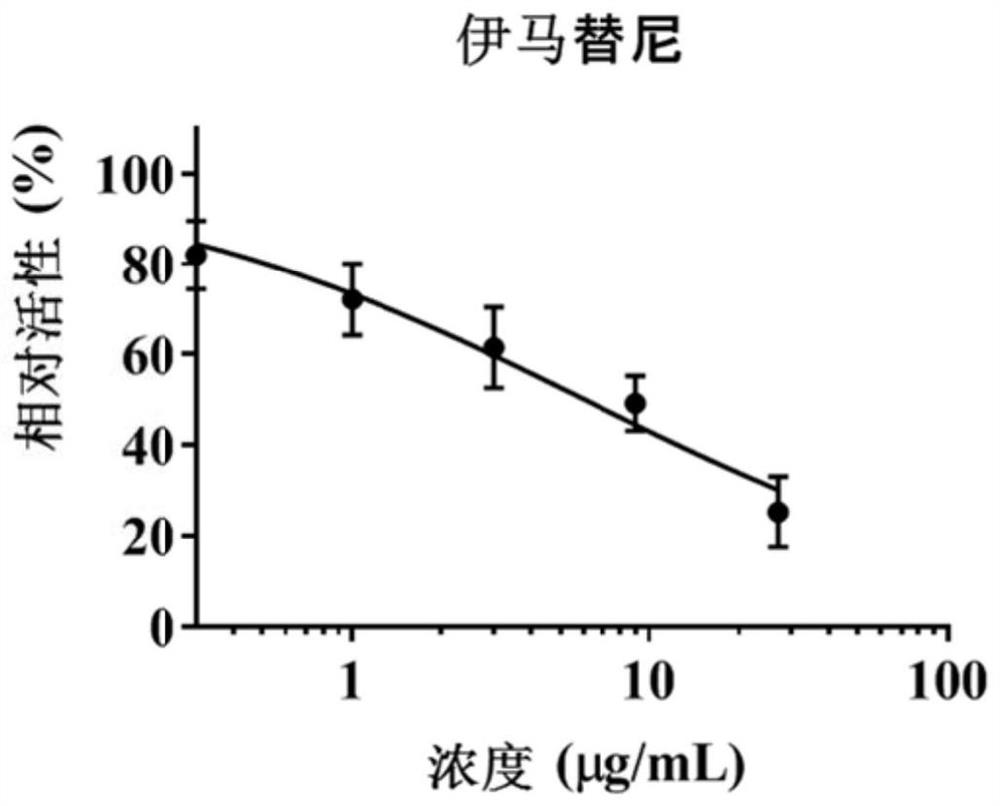
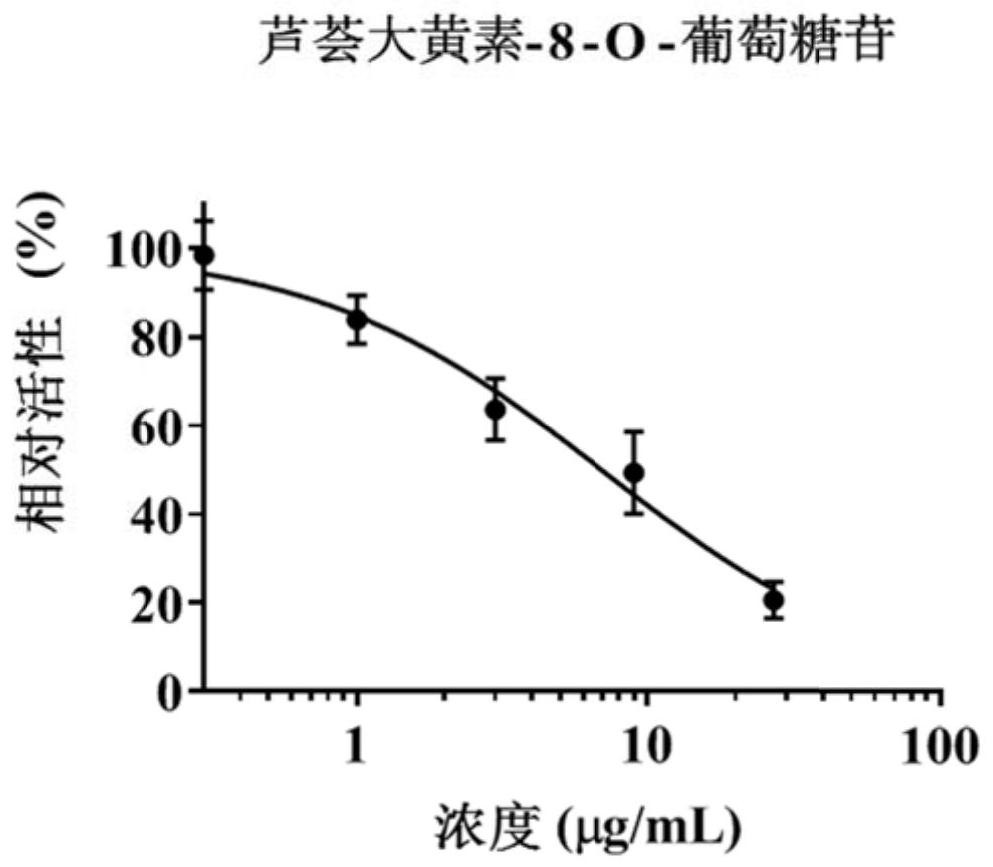
[0084] Example 2 Hepatotoxicity investigation
[0085] In order to further verify the accuracy of the method in Example 1 and the correlation between MRP2 / 3 inhibition and liver toxicity, this example conducted a hepatotoxicity investigation.
[0086] HepaRG cells in the logarithmic growth phase were digested and adjusted to 5×10 4 cells / mL, inoculate about 5000 cells per well in a 96-well plate, and administer after incubation for 18-24 hours. A blank control group (without HepaRG cells), a vehicle control group (0.5% DMSO), a positive control group (MRP2 / MRP3 inhibitors) and different concentrations of target compound administration groups were set up. Firstly, a preliminary experiment was carried out according to the solubility of the compound in the solvent, and the concentration range of each target compound was preliminarily determined, and then further toxicity tests were carried out according to the results of the preliminary experiment to determine the half-inhibitor...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com