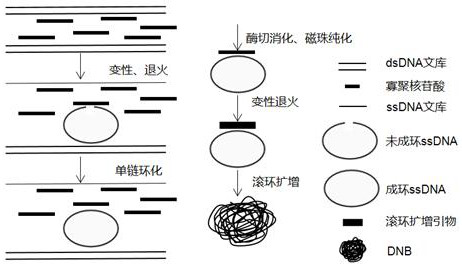
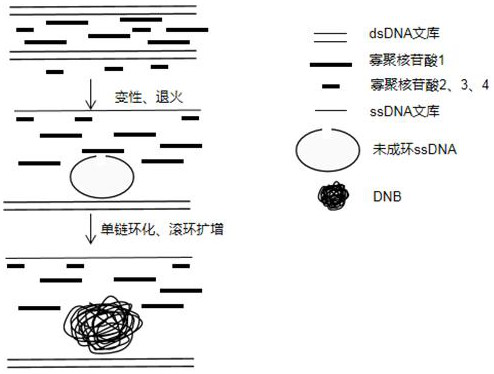
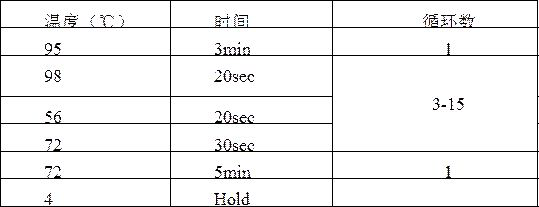
A kit and method for constructing DNA nanospheres in one step
A nanosphere and kit technology, applied in the field of high-throughput sequencing, can solve the problems of many processes and long time, and achieve the effect of saving operation time, saving operation steps, and improving the efficiency of circularization
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0052] Using a batch of actual samples submitted for clinical metagenomic sequencing (mNGS) as experimental materials, first extract the nucleic acid (DNA and / or RNA) of the samples, then build a library, and finally construct a DNB that can be used for sequencing on the MGI platform.
[0053] The specific experimental steps are as follows:
[0054] 1. Sample nucleic acid extraction: The DNA extraction kits used in this example are the Microbial Genomic DNA Extraction Kit (Cat. No. PMD101) from Nanjing Practice Medical Laboratory Co., Ltd., and the Microbial Genomic DNA Extraction Kit from Sputum / Nasal Secretion Samples. (Cat. No. PMD102) and Nasopharyngeal Swab / Anal Swab Sample Microorganism Genomic DNA Extraction Kit (Cat. No. PMD103); Pharyngeal swab / anal swab sample microbial RNA extraction kit (Cat. No. PMD203).
[0055] 1.1 The specific experimental process of DNA extraction is as follows:
[0056] (1) Sample processing:
[0057] A. Blood: Centrifuge at 2100rpm for 5m...
Embodiment 2
[0208] Sample: Another batch of actual samples submitted for metagenomic sequencing (mNGS) for clinical examination.
[0209] The difference from Example 1 is that the DNB preparation buffer components in the DNB preparation process are 80mM potassium acetate, 20mM magnesium acetate, 50mM Tris-acetic acid, 0.3mg / mlBSA, 10mMDTT, 5μM oligonucleotide 1, 5μM oligonucleotide Nucleotide 2, 1 μM oligonucleotide 3, 5 μM oligonucleotide 4;
[0210] DNB polymerase buffer components are 10mM potassium acetate, 20mM magnesium acetate, 40mM Tris-acetic acid, 0.3mg / mlBSA, 5mMDTT, 5mMATP and 5mMdNTP;
[0211] The working concentration of each component of the DNB polymerase mixture is 1U / μl Phi29DNAPolymerase, 0.5U / μl T4DNALigase, 50ng / μl pyrophosphatase, 0.5U / mlT4Gene32Protein;
[0212] EDTA concentration in DNB stop buffer is 500mM;
[0213] sample number sample type nucleic acid type Nucleic acid concentration ng / μl Library concentration ng / μl total library ng 1 ...
Embodiment 3
[0219] Sample: Another batch of actual samples submitted for metagenomic sequencing (mNGS) for clinical examination.
[0220] Different from Example 1 and Example 2, the DNB preparation buffer components in the DNB preparation process are 60mM potassium acetate, 15mM magnesium acetate, 30mM Tris-acetic acid, 0.2mg / mlBSA, 4mMDTT, 2.5μM oligonucleotide 1 , 2 μM oligonucleotide 2, 0.03 μM oligonucleotide 3, 2 μM oligonucleotide 4;
[0221] DNB polymerase buffer components are 5mM potassium acetate, 10mM magnesium acetate, 20mM Tris-acetic acid, 0.2mg / mlBSA, 4mMDTT, 2mMATP and 2mMdNTP;
[0222] The working concentration of each component of the DNB polymerase mixture is 0.2U / μl Phi29DNAPolymerase, 0.3U / μl T4DNALigase, 10ng / μl pyrophosphatase, 0.2U / mlT4Gene32Protein and 0.2U / mlE.coliSSB.
[0223] The concentration of EDTA in DNB stop buffer is 250mM;
[0224] sample number sample type nucleic acid type Nucleic acid concentration ng / μl Library concentration ng / μl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com