Kit and special primer for detecting GI.1 type norovirus in clinical sample
A GI.1, kit technology, applied in the field of biology, can solve the problems of high cost and long time consumption
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Embodiment 1, acquisition of special primers for detection of GI.1 type norovirus
[0037] A variety of primers were designed after a large number of sequence analysis, and the performance of the primers was verified through preliminary experiments, and finally the primer pair with the best performance for identifying GI.1 Norovirus was obtained.
[0038] Primer pair I is as follows (target sequence is 116bp):
[0039] Upstream primer F1 (sequence 1): 5'-TTGTAGTTGCAGGGCGAGTT-3';
[0040] Downstream primer R1 (SEQ ID NO: 2): 5'-CAATGGCAGATTTGGGAGTG-3'.
[0041] Primer pair II is as follows (target sequence is 209bp):
[0042] Upstream primer F2 (sequence 3): 5'-GACCTGCCCTAGTCCTGATT-3';
[0043] Downstream primer R2 (SEQ ID NO: 4): 5'-CCAAACGACCGTCCAAAGTA-3'.
[0044] Each primer was artificially synthesized separately.
Embodiment 2
[0045] Embodiment 2, application primer pair detection positive plasmid
[0046] 1. Preparation of positive plasmid
[0047] Positive plasmid I is a circular plasmid, as shown in sequence 5 of the sequence listing.
[0048] Positive plasmid II is a circular plasmid, as shown in sequence 6 of the sequence listing.
[0049] 2. Application of primer pairs to detect positive plasmids
[0050] 1. Dilute positive plasmid I with double distilled water to obtain template solution I; in template solution I, the concentration of positive plasmid I is 4.105×10 8 / μL. Dilute positive plasmid II with double distilled water to obtain template solution II; in template solution II, the concentration of positive plasmid II is 3.94×10 8 copies / μL.
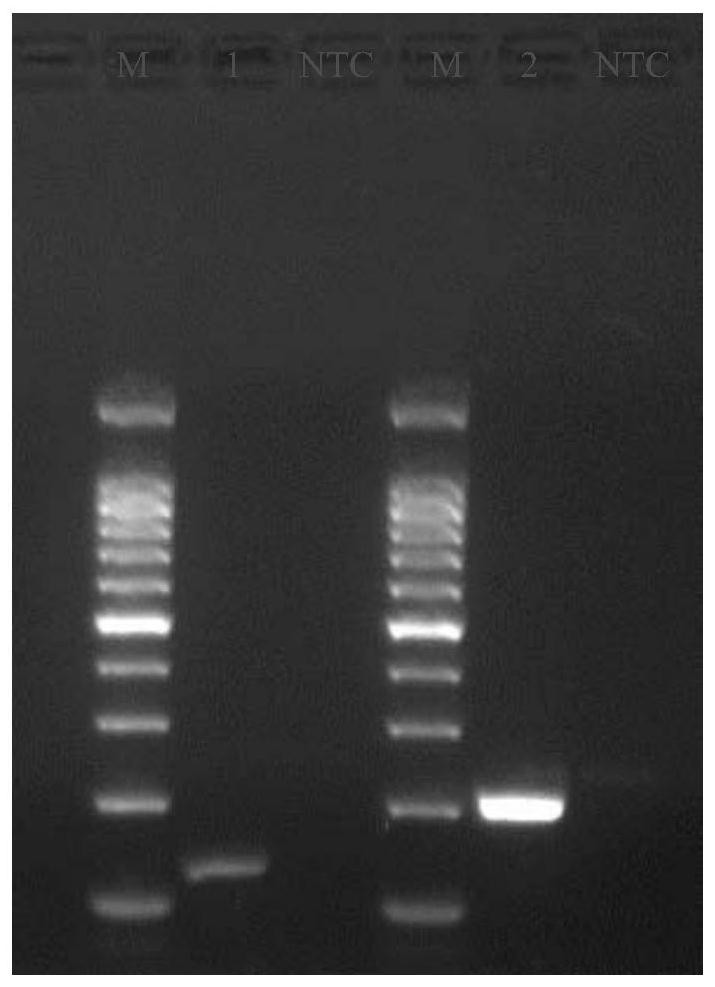
[0051] 2. Take the template solution, carry out PCR amplification, and then carry out 1.5% agarose gel electrophoresis.
[0052]PCR amplification reaction system (25 μL): DreamTaq Green PCR Master Mix (2X) 12.5 μL, upstream primer 1 μL, downst...
Embodiment 3
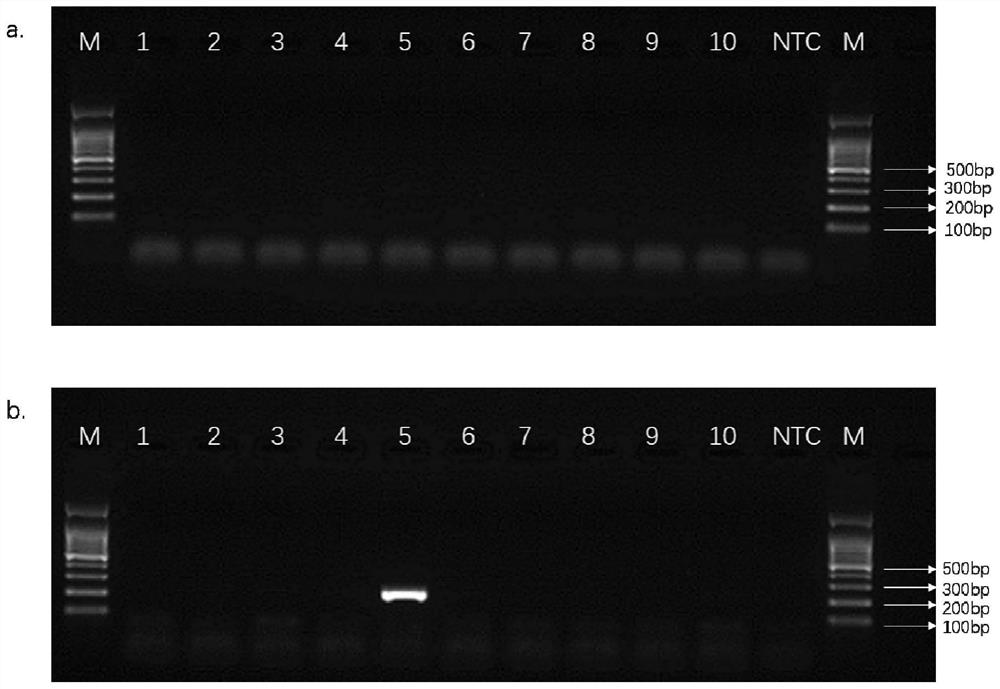
[0066] Embodiment 3, the detection of stool sample GI.1 type norovirus
[0067] Human stool samples: Samples 1-5 are norovirus-positive human stool samples from the intestinal tract clinic of Haidian Hospital; samples 6-10 are norovirus-negative human stool samples from the intestinal tract clinic of Haidian Hospital.
[0068] 1. Preparation of template solution
[0069] Human feces samples were taken, total RNA was extracted using a kit (QIAGEN RNeasyPower Microbiome Kit), and cDNA was obtained by reverse transcription. The cDNA is in the form of a solution, which is the template solution.
[0070] 2. Conduct detection
[0071] Take the template solution, use primer pair I or primer pair II to carry out PCR amplification respectively, and then carry out 1.5% agarose gel electrophoresis.
[0072] PCR amplification reaction system (25 μL): DreamTaq Green PCR Master Mix (2X) 12.5 μL, upstream primer 1 μL, downstream primer 1 μL, template solution 1 μL, sterile water 9.5 μL. ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com