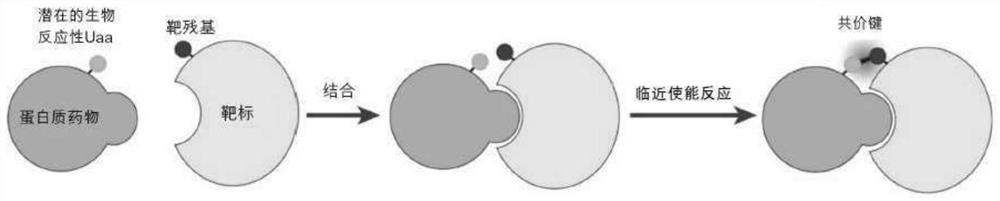
Covalent protein drugs developed via proximity-enabled reactive therapeutics (PERX)
A protein and drug technology, applied in the field of covalent protein drugs developed through proximity-enabling response therapy, which can solve the problems of lack of covalent reactivity of natural amino acid residues, selective application of reactions, obstacles, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
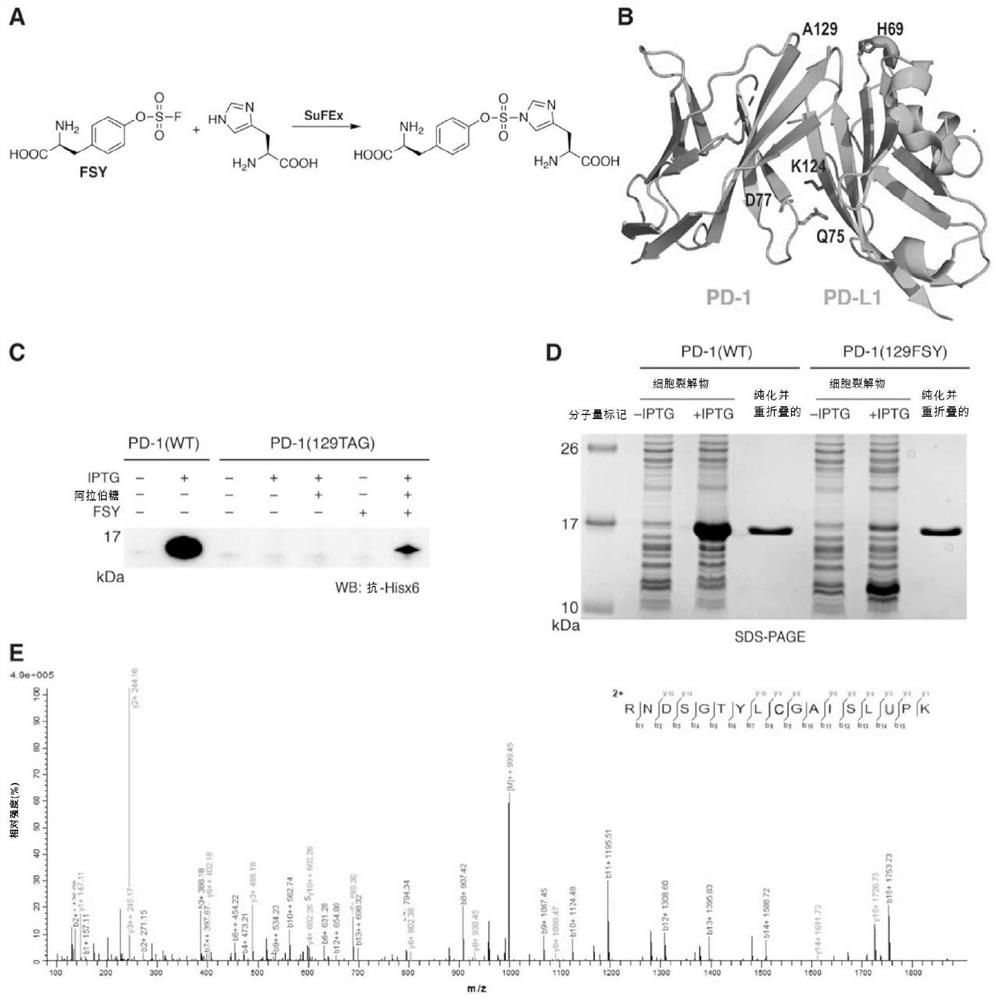
[0108] Cloning of human PD-1 and PD-L1 extracellular IgV domains
[0109] Protein stability aided by replacement of human PD-1 (hPD-1) IgV domain encoding gene (residues 32-160, Cys93 with Ser (C93S) 28 ) into the pET-26b vector with a carboxy-terminal 6x histidine tag to generate plasmid pET-26b-PD1(WT) (SEQ ID NO: 15 and 22). Briefly, plasmid pUC57-Kan-hPD-1 IgV domain (32 -160, C93S) to amplify the DNA of the hPD-1 IgV domain (32-160, C93S), which was codon-optimized and synthesized by GENEWIZ (China, Suzhou). The PCR product was digested with NdeI-HF and XhoI-HF and ligated into the pre-cut pET-26b vector using T4 DNA ligase (NEB, catalog number M0202L).
[0110] PD-1 WT-F:GTTAGACTcatatgTGGAATCCGCCGACCTTTAGC (SEQ ID NO:1)
[0111] PD-1 WT-R: ACTGctcgagCGGACTAGGACTCGGATGTGCG (SEQ ID NO: 2)
[0112] To introduce a TAG codon at site D77 or A129, overlap PCR was performed using Q5 high-fidelity DNA polymerase and using plasmid pET-26b-PD1(WT) as a template using the follow...
Embodiment 2
[0130] Expression and purification of human PD-1 IgV domain and PD-L1 IgV domain
[0131] Plasmids pET-26b-PD1(WT), pET-26b-PD-L1(WT) and pET-26b-PD-L1(H69A) were individually transformed into E. coli BL21(DE3) electrocompetent cells. Plasmid pET-26b-PD-1(Q75TAG), pET-26b-PD-1(D77TAG) or pET-26b-PD-1(A129TAG) each with plasmid pEvol-FSYRS 29 Co-transformation into Escherichia coli BL21 (DE3) electrocompetent cells.
[0132] For the expression of PD-1(WT), PD-L1(WT) and PD-L1(H69A), the transformed bacteria were cultured at 37°C in 2xYT medium containing 50 μg / mL kanamycin, and when OD 600 Induction with 1 mM IPTG when reaching 0.8. For FSY incorporation into proteins, the transformed bacteria were cultured at 37°C in 2xYT medium containing 50 μg / mL kanamycin and 34 μg / mL chloramphenicol, and when OD 600 At 0.8 it was induced with 1 mM IPTG, 0.2% arabinose and 1 mM FSY.
[0133]After induction of expression for 12 hours, bacteria were harvested by centrifugation at 7000 rpm...
Embodiment 3
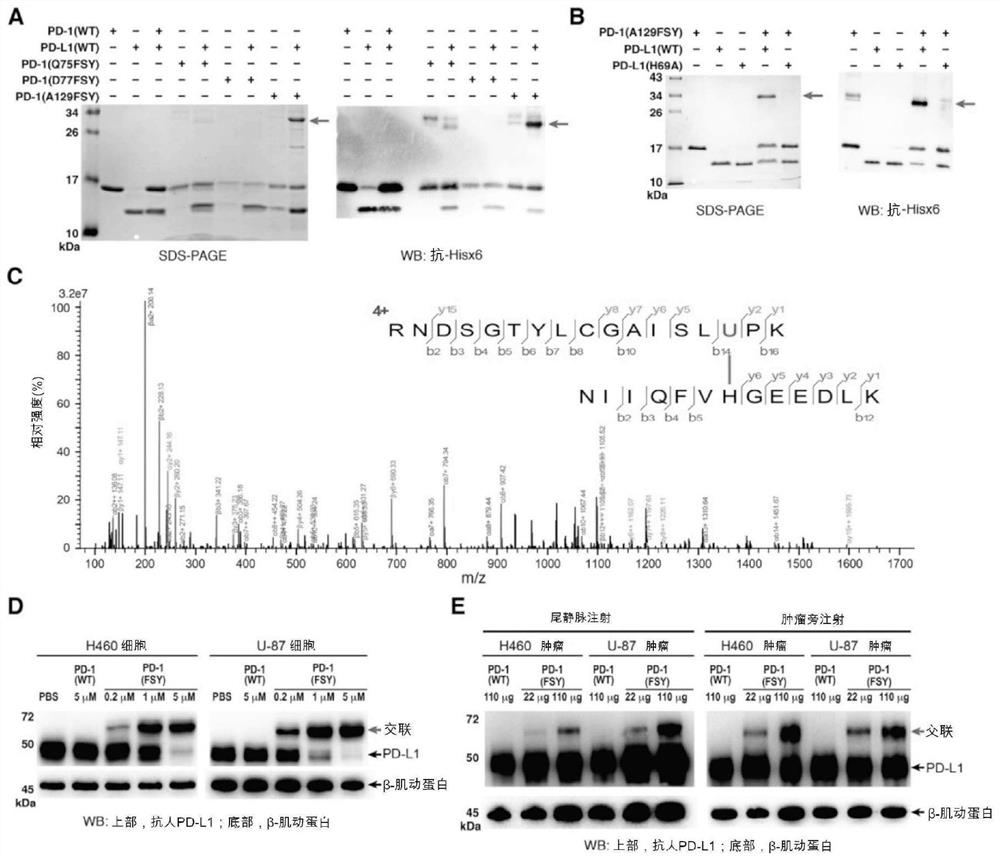
[0140] Cross-linking of PD-1(FSY) and PD-L1
[0141] 3.1 In vitro cross-linking of PD-1(FSY) and PD-L1
[0142] Purified and refolded PD-1(WT), PD-1(Q75FSY), PD-1(D77FSY) or PD-1(A129FSY) and PD-L1(WT) or PD-L1(H69A) at a ratio of 1:1 The molar ratios were incubated in PBS buffer at 37 °C for 6 h. The amount of PD-L1 was 4 μg. After incubation, 5x reducing loading buffer (CWBio, cat# CW0027) was added to the incubation and heated at 100°C for 10 min. These samples were then separated by 15% SDS-PAGE gels followed by Coomassie blue staining. Then use a primary antibody specific for the His6x tag (mAb anti-6xHis tag, Abcam, Cat. No. 18184, 1:1000 dilution) and a secondary antibody goat pAb anti-mouse IgG (Abcam, Cat. No. 97023, 1:5000 dilution) Perform western blotting. Protein bands were visualized by chemiluminescence (Bio-rad, catalog #1705062).
[0143] 3.2 Mass spectrometry analysis
[0144] PD-1(FSY) and PD-1(FSY) / PD-L1 cross-linked protein samples were digested wit...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com