Polypeptide with binding affinity to HPV16E6 protein and application thereof
A HPV16E6, affinity technology, applied in the field of biomedicine, can solve the problems of serious immunogenicity, toxic and side effects, reduced function, high cost, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0083] Example 1 Wencative Construction and Screening of HPV16E6 binding polypeptide
[0084] The phage showing the random combination library of HPV16E6 binding polypeptides, that is, many different SPA domain-related polypeptides, HPV16E6 binding polypeptides, and identified their affinity.
[0085] 1. Construction and identification of the random combination phage display library of HPV16E6 binding polypeptide
[0086] According to the amino acid sequence and structure of wild type SPA-Z (Nilsson B et al, Protein ENG.1987; 1 (2): 107-113), a random primer is designed for the coding sequence corresponding to the three helix structural regions, and the PCR method is expanded. Increasing the SPA coding sequence that can cause random amino acid mutations, named SPA-N.
[0087] 2, construction of PCANTAB5E / SPA-N recombinant plasmid
[0088] Choose the expression of affinity in the M13 phage system (purchased from Beijing Baocovi Food Anni Company), with Pcantab5e (purchased from B...
Embodiment 2
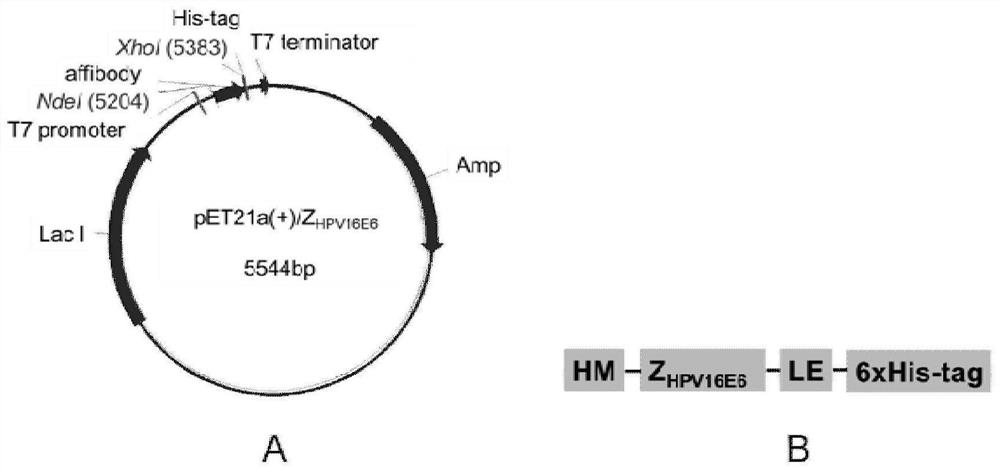
[0095] Example 2 HPV16E6 binding polypeptide recombinant plasmid structure and prokaryotic expression and purification
[0096] Select 3 clones with higher PHAGE-ELISA reading as before, figure 1 Zeng HPV16E6 1115, Z HPV16E6 1171, Z HPV16E6 1235) In order to perform functional detection of the screening of Affibody molecules, the recombinant plasmid is built, the expression of the prototype protein and its identification, and purified protein are prepared.
[0097] 1. Recombinant plasmid construction and identification of PET21A (+) / Affibody
[0098] Refer to the Affibody gene sequence (GenBank: GY324633.1) Design PCR primer, upstream primers 5'GGAATTC Catatg GTTGACAACAAATTCAACAAAAAGAA 3 '(SEQ ID NO: 10, the underscore represents NED I enzyme dug point), downstream primer 5'ccg CTCGAG TTTCGGAGCCTGAGCGTCG3 '(SEQ ID NO: 11, the underscore represents an XHO I enzyme dug bit); the sequence of sequencing is correct, the three-level library single cloned AFFIBODY Z HPV16E6 1115, Z HPV...
Embodiment 3
[0101] Example 3, Z HPV16E6 Affibody polypeptide combination with HPV16E6 protein
[0102] To identify Z HPV16E6 Affibody polypeptide and HPV16E6 protein-binding specificity, three Zs, screening, plasma resonance techniques (SPR) HPV16E6 1115, Z HPV16E6 1171, Z HPV16E6 1235 molecules and their control ZwtaffiBody and the specific binding of target protein HPV16E6.
[0103] (1) Preparation and identification of HPV16E6 recombinant proteins
[0104] The HPV16E6 gene was amplified from the PCR method, cloned to the PET21a (+) vector, and constructed PET21A (+) / HPV16E6 recombinant plasmid, and transformed sequencing identification and completely correct recombinant plasmid PET21a (+) / HPV16E6 to E.COLI BL21 (DE3) strain. Inducing expression by 1 mm IPTG, SDS-PAGE electrophoresis showed more concentrated induced strips at 14kda, consistent with the theoretical value ( Figure 5 ). After ultrasonic cracking, SDS-PAGE electrophoresis analyzes recombinant proteins in the precipitate in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com