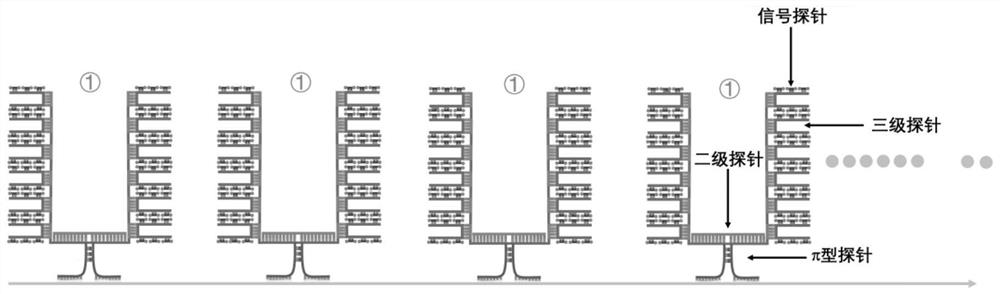
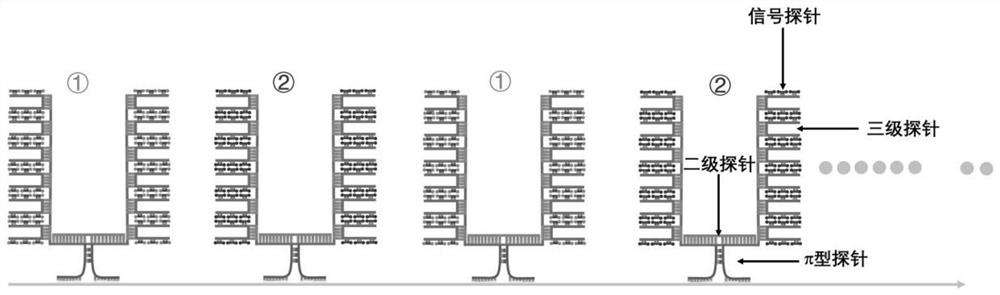
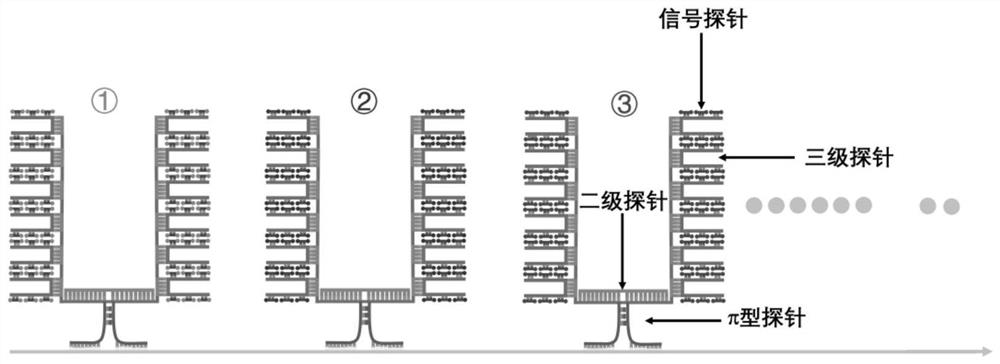
π-fish localized single-molecule probe composition and its application in nucleic acid in situ detection
An in-situ detection and composition technology, which is applied in the direction of microbial determination/inspection, recombinant DNA technology, biochemical equipment and methods, etc., can solve the problems of limited signal amplification ability and cost increase, and achieves non-degradable and low background noise , the effect of high signal specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0060] like figure 1 The single gene detection shown is a method for in situ detection of Actb gene mRNA and DNA in BHK cells by π-FISH. The specific steps are as follows:
[0061] (1) Probe design:
[0062] a. Primary π-type probes: 17 primary π-type probe pairs were designed for the Actb gene in BHK cells, and each primary π-type probe includes a π foot region, a π top region and a π middle region; ,
[0063] The π foot region is the specific hybridization region that is complementary to the base pairing of Actb mRNA, which is represented by NNN… The complementary sequences of the regions are AAGTCCTT and TTCCACTA respectively;
[0064] The sequence on the left side of the primary π-type probe: 5'-NNN...NNNAAGTCCTTNNN...NNN-3'
[0065] The sequence on the right side of the primary π-type probe: 5'-NNN...NNNTTCCACTANNN...NNN-3'
[0066] b. Secondary probe: The secondary probe is divided into a middle region and two end regions; the middle region is a sequence complementa...
Embodiment 2
[0114] like Figures 7 to 8 The single-gene and multi-gene detection in the shown tissue sections, namely, π-FISH in situ detection of Cux2 gene, Pcp4 gene and co-detection of the mRNA of Pcp4 and Rorb genes in mouse brain tissue sections, the specific steps are as follows:
[0115] (1) Probe design:
[0116] a. Primary π-type probes: The design principles of π-type probes for Cux2, Pcp4 and Rorb genes are the same as those in Example 1 a; 19 π-type probes are designed for Cux2 gene, and 8 π-type probes are designed for Pcp4 gene, Rorb gene designed 17 π-type probes, each of which is divided into three regions, namely the π foot region, the π top region and the π middle region. The pi foot region is the specific hybridization region that is complementary to the base pairing of Cux2, Pcp4 and Rorb gene mRNAs, and is represented by NNN... The sequence of the π-type π top region is represented by NNN...
[0117] The sequence information of the primary π-type probe of Cux2 and ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| thickness | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com