Toxic protein gene leakage expression cloning method
A cloning method, a technology of toxic proteins, applied in the biological field, can solve problems such as low copy number, protein leakage expression, host bacteria death, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] A method for cloning expression due to the leakage of toxic protein genes, comprising the following steps:
[0030] Step S1, adding the SsrA polypeptide sequence to the C-terminus of the CcdB protein, the sequence is as follows:
[0031] atgcagtttaaggtttacacctataaaagagagagccgttatcgtctgtttgtggatgtacagagtgatatcattgacacgcccgggcgacggatggtgatccccctggccagtgcacgtctgctgtcagataaagtctcccgtgaactttacccggtggtgcatatcggggatgaaagctggcgcatgatgaccaccgatatggccagtgtgccggtgtccgttatcggggaagaagtggctgatctcagccaccgcgaaaatgacatcaaaaacgccattaacctgatgttctggggaataGCTGCTAACGACGAAAACTACGCTCTGGCTGCTTAA;
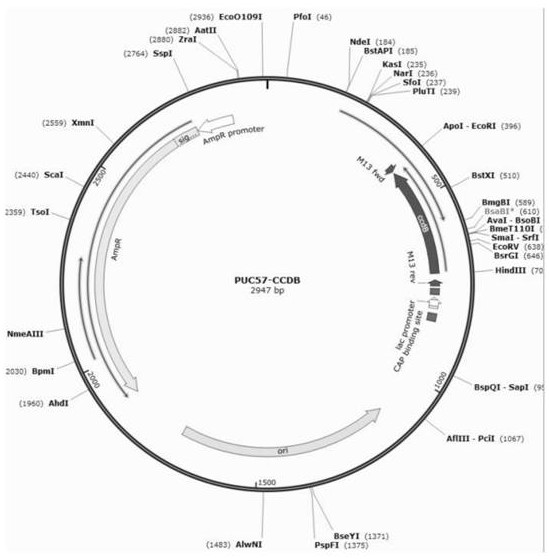
[0032] Step S2, obtain the gene fragment by PCR amplification method, and then use Gibson to recombine into the target vector PUC57 to construct the complete sequence, such as figure 1 shown;
[0033] Step S3. After connecting and cloning the target gene sequence into the expression vector, transform Escherichia coli, culture for 12-16 hours, observe the condition of spots and perform PCR colony scr...
Embodiment 2
[0042] A method for cloning expression due to the leakage of toxic protein genes, comprising the following steps:
[0043] Step S1, adding the SsrA polypeptide sequence to the C-terminus of the CcdB protein, the sequence is as follows:
[0044] atgcagtttaaggtttacacctataaaagagagagccgttatcgtctgtttgtggatgtacagagtgatatcattgacacgcccgggcgacggatggtgatccccctggccagtgcacgtctgctgtcagataaagtctcccgtgaactttacccggtggtgcatatcggggatgaaagctggcgcatgatgaccaccgatatggccagtgtgccggtgtccgttatcggggaagaagtggctgatctcagccaccgcgaaaatgacatcaaaaacgccattaacctgatgttctggggaataGCTGCTAACGACGAAAACTACGCTCTGGCTGCTTAA;
[0045] Step S2, obtain the gene fragment by PCR amplification method, and then use Gibson to recombine into the target vector PUC57 to construct the complete sequence;
[0046] Step S3: After connecting and cloning the target gene sequence into the expression vector, transform Escherichia coli, culture for 12-16 hours, observe the condition of spots and perform PCR colony screening for positive clone...
Embodiment 3
[0055] A method for cloning expression due to the leakage of toxic protein genes, comprising the following steps:
[0056] Step S1, adding the SsrA polypeptide sequence to the C-terminus of the CcdB protein, the sequence is as follows:
[0057] atgcagtttaaggtttacacctataaaagagagagccgttatcgtctgtttgtggatgtacagagtgatatcattgacacgcccgggcgacggatggtgatccccctggccagtgcacgtctgctgtcagataaagtctcccgtgaactttacccggtggtgcatatcggggatgaaagctggcgcatgatgaccaccgatatggccagtgtgccggtgtccgttatcggggaagaagtggctgatctcagccaccgcgaaaatgacatcaaaaacgccattaacctgatgttctggggaataGCTGCTAACGACGAAAACTACGCTCTGGCTGCTTAA;
[0058] Step S2, obtain the gene fragment by PCR amplification method, and then use Gibson to recombine into the target vector PUC57 to construct the complete sequence;
[0059] Step S3: After connecting and cloning the target gene sequence into the expression vector, transform Escherichia coli, culture for 12-16 hours, observe the condition of spots and perform PCR colony screening for positive clone...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com