Method and kit for constructing multi-gene mutation sequencing library of lung cancer driving genes
A technology for driving genes and sequencing libraries, applied in the field of biomedicine, can solve the problems of high cost, cumbersome and complicated operation, and long time.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
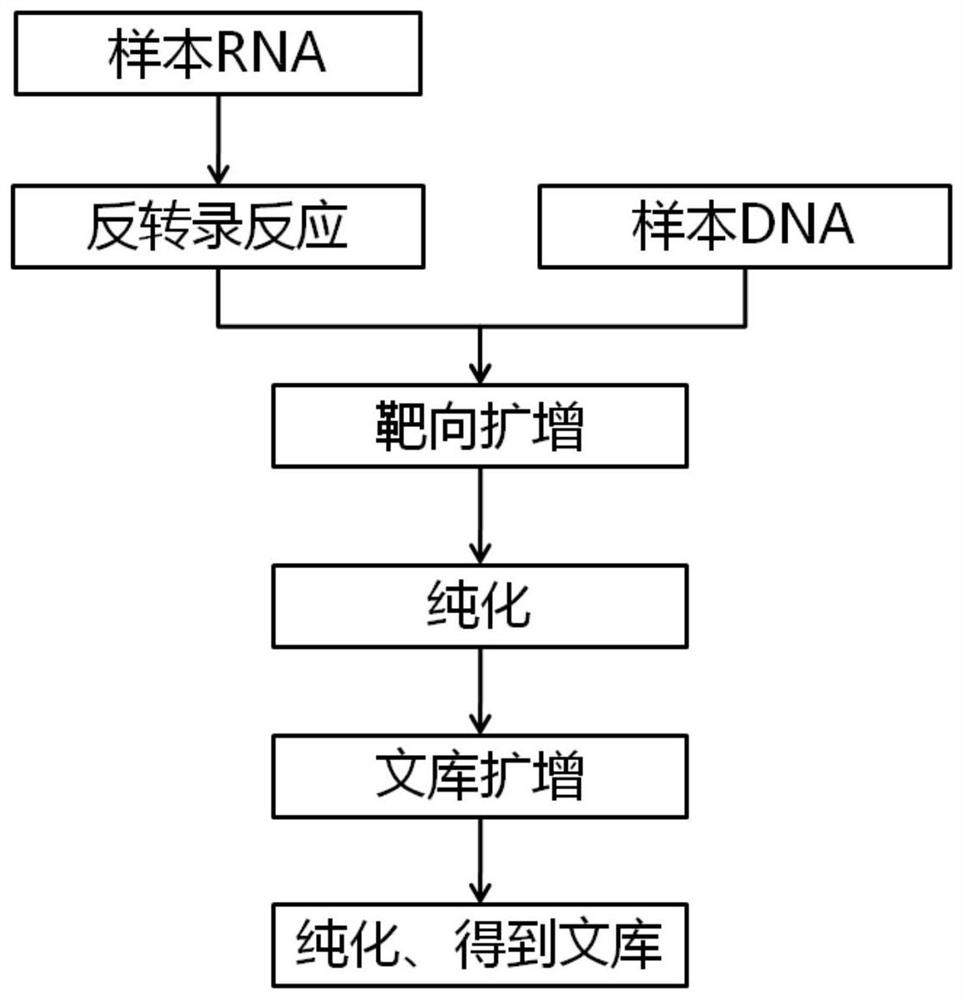
[0075] A kit for detecting multi-gene somatic mutation and gene fusion of lung cancer, the kit includes reverse transcription buffer, targeted amplification buffer, library amplification buffer, forward library amplification primer A5I, reverse library amplification Add primer A7I, purified magnetic beads, eluent, quality control.
[0076] Reverse transcription buffer, including reverse transcriptase, dNTP, buffer, random primer or Oligo(dT) primer, RNase inhibitor, used to reverse transcribe sample RNA into cDNA.
[0077] Targeted amplification buffer, including high-fidelity DNA polymerase, PCR buffer, dNTP mixture, and targeted amplification primers, are used to amplify the target mutation region in the nucleic acid to be tested. The target amplification primers (SEQ ID NO.1-SEQ ID NO.89), each primer structure includes a universal primer sequence and a specific primer sequence, and the amplification region at least covers all target gene mutation sites to be detected.
[...
Embodiment 2
[0094] Sample detection was performed using the kit described in Example 1.
[0095] 1. Sample DNA and RNA extraction:
[0096] Fresh tissue or cells, paraffin tissue samples, etc. were used as samples. DNA and RNA were extracted using a commercial company’s DNA / RNA extraction kit according to the instructions, and the concentration was measured with a Qubit4 fluorometer.
[0097] The loading amount of total RNA should not be lower than 200ng, and the loading amount of total DNA should not be lower than 50ng.
[0098] 2. Reverse transcription reaction: For each sample to be tested, configure the reaction system proportionally in a 0.2 μl PCR tube as shown in Table 3:
[0099] Table 3 reverse transcription system
[0100] Reagent name The actual amount sample total RNA 200-1000ng reverse transcription buffer 2μl nuclease free water Make up to 10 μL
[0101] After the system configuration is completed, use a pipette to mix gently by pipett...
Embodiment 3
[0132] Example 3 Effects of different sample nucleic acid concentrations on library construction
[0133]Select 5 examples of samples with different mutation types verified by Sanger sequencing, use the kit of Example 1, and according to the method of Example 2, add 10ng, 30ng, 50ng, 100ng, and 200ng sample DNA to carry out library construction, the results show (as shown in Table 10), when the nucleic acid concentration of the sample for constructing the library is as low as 10 ng, the corresponding mutation can still be detected correctly using the library construction kit and construction method provided by the present invention, and the results of the library construction using different initial amounts of DNA are measured. The mutation rate is close.
[0134] Table 10 Test results of sample library construction with different nucleic acid concentrations
[0135]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com