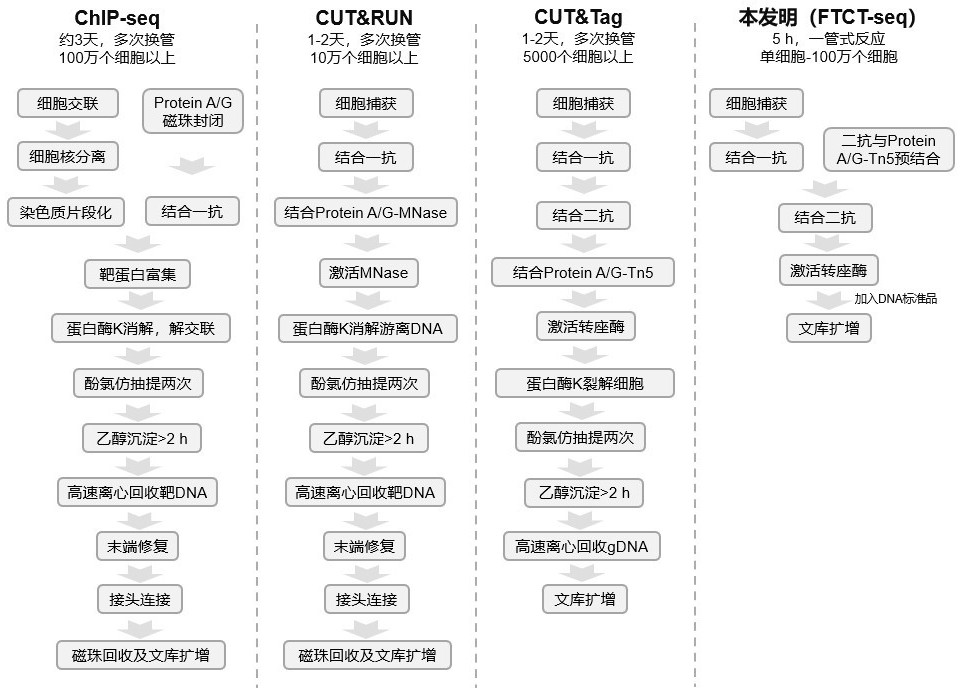
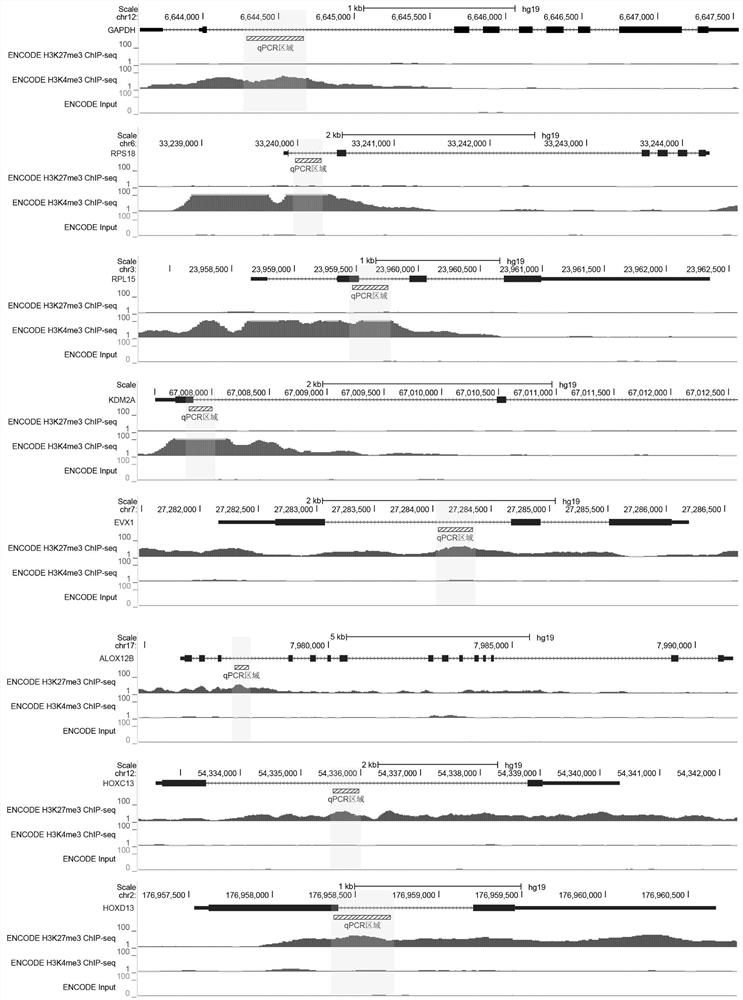
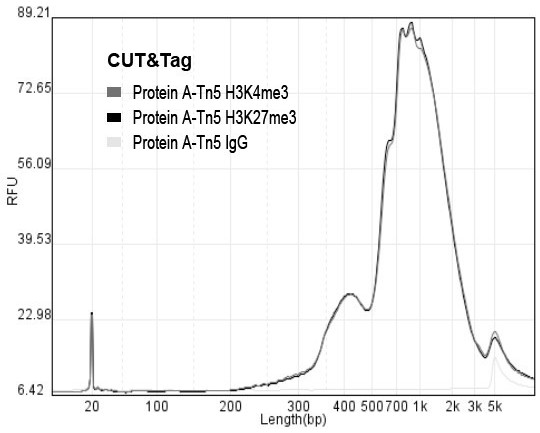
Rapid library building method for identifying target protein chromatin binding map
A technology for chromatin and target identification, which is applied in the field of rapid library building for identifying chromatin binding maps of target proteins.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0090]Example 1: The effect of secondary antibody and recombinant fusion transposase Protein A-TN5 and Protein Ag-TN5 pre-bonded to library yield.
[0091]Process diagramFigure 7 The specific process is as follows:
[0092]1. Collect cells. Take about 100,000 293FT cells, and the cells were centrifuged at room temperature for 600 x g. It was cleaned twice with a cleaning buffer (20 mm hepes (pH 7.5), 150 mm NaCl, 0.5 mm, 1x protease inhibitorscocktail). The cells were resuspended in 90 μL of cleaning buffer.
[0093]2. Activate magnetic beads and capture cells. 10 μl of CONA magnetic beads were added to the binding buffer (20 mM HEPES (pH 7.5), 10 mM KCl, 1 mM CaCl2, 1 mm MnCl2) washed twice. The magnetic beads were hung in 10 μl binding buffer. The magnetic beads were added to the cells, and the room temperature was 10 to 10 min.
[0094]3. Anti-combination. Three kinds of dilutions were diluted in 1: 100, using an anti-dilution buffer (20 mm hepes (pH 7.5), 150 mm NaCl, 0.5 mm amine, 0.05% Di...
Embodiment 2
[0109]Example 2: The effect of different genomic DNA extraction methods on library yield.
[0110]Using recombinant fusion transposase Protein Ag-TN5 and H3K27ME3 antibody to verify the effect of different genomic DNA extraction methods on library production, flow schematicFigure 12 The specific process is as follows:
[0111]1. Collect cells. Take about 100,000 293FT cells, and the cells were centrifuged at room temperature for 600 x g. It was cleaned twice with a cleaning buffer (20 mm hepes (pH 7.5), 150 mm NaCl, 0.5 mm, 1x protease inhibitorscocktail). The cells were resuspended in 90 μL of cleaning buffer.
[0112]2. Activate magnetic beads and capture cells. 10 μl of CONA magnetic beads were added to the binding buffer (20 mM HEPES (pH 7.5), 10 mM KCl, 1 mM CaCl2, 1 mm MnCl2) washed twice. The magnetic beads were hung in 10 μl binding buffer. The magnetic beads were added to the cells, and the room temperature was 10 to 10 min.
[0113]3. Anti-combination. Two kinds of dilutions were dilu...
Embodiment 3
[0133]Example 3: Protein A-TN5 and Protein AG-TN5 in FTCT-SEQ buildings.
[0134]According to the improvement of Examples 1 and Example 2, we invented a rapid settlement method for identifying a target protein DNA binding map and named FTCT-SEQ. The reorganized fusion transposase Protein A-TN5 and Protein AG-TN5 verify the construction of the method of the process of the invention, the specific process is as follows:
[0135]1. Collect cells. Take about 100,000 293FT cells, and the cells were centrifuged at room temperature for 600 x g. It was cleaned twice with a cleaning buffer (20 mm hepes (pH 7.5), 150 mm NaCl, 0.5 mm, 1x protease inhibitorscocktail). The cells were resuspended in 90 μL of cleaning buffer.
[0136]2. Activate magnetic beads and capture cells. 10 μl of CONA magnetic beads were added to the binding buffer (20 mM HEPES (pH 7.5), 10 mM KCl, 1 mM CaCl2, 1 mm MnCl2) washed twice. The magnetic beads were hung in 10 μl binding buffer. The magnetic beads were added to the cells, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com