Cellulosome with improved catalytic activity and assembly method and application thereof
A technology for improving catalytic activity and cellulosomes, applied in chemical instruments and methods, methods based on microorganisms, biochemical equipment and methods, etc., can solve the problem of low catalytic activity of artificial cellulosomes, and achieve synergistic catalytic effect, The effect of reducing consumption and production cost and good application prospect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Fusion of Lignocellulose Degrading Enzyme Gene and Docking Protein Gene
[0037] The coding genes of docking proteins Doc1 and Doc2 were derived from R. flavefaciens (GenBank: WP_009985128) and R. flavefaciens (GenBank: 5M2O-B), respectively, and the nucleotide sequences were optimized according to the codon preference of E. coli, Doc1 and Doc2 The nucleotide sequences of Doc1 and Doc2 are shown in SEQ ID NO.1 and SEQ ID NO.2, and the amino acid sequences of Doc1 and Doc2 are shown in SEQ ID NO.6 and SEQ ID NO.7.
[0038] The coding genes of Doc1 and Doc2 were respectively fused to the C genes of xylanase AExynM (GenBank: HQ724284.1) and glucanase EG1 (GenBank: AF329732) with high catalytic activity preserved in the laboratory by overlapping PCR. At the end, the primer sequences are shown in Table 1, and the specific steps are as follows:
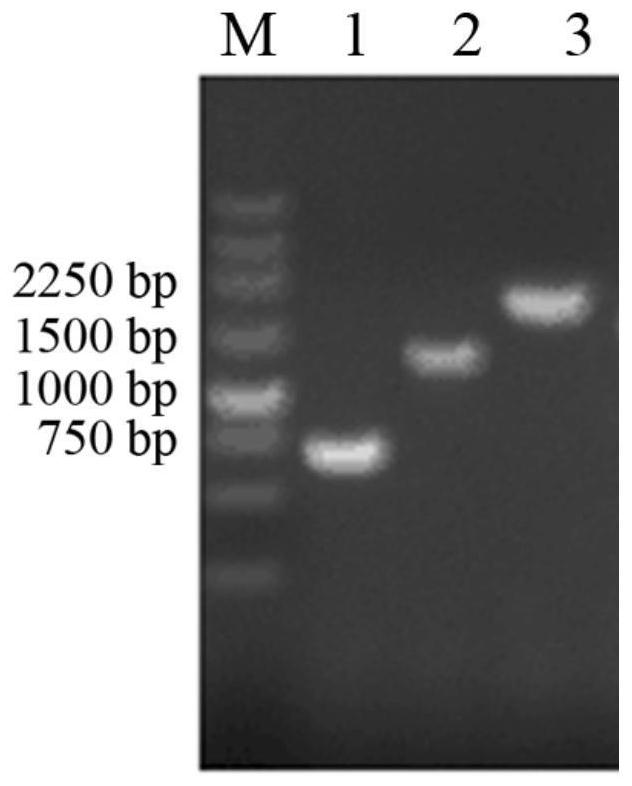
[0039] Use the pET-32a(+)-AExynM preserved in this experiment as a template, and use Xyn-F and Xyn-R as primers for the first roun...
Embodiment 2
[0044] Induced expression of fusion genes
[0045] Correctly sequenced Escherichia coli recombinants pET-32a(+)-AExynM-Doc1 and pET-32a(+)-EG1-Doc2 and BL21-pET-32a(+)-AExynM and BL21-pET-32a preserved in the laboratory (+)-EG1 was subjected to IPTG induction culture respectively: inoculate the strains in LB medium respectively, cultivate overnight as the seed medium, inoculate 1% into 30mL LB medium, and cultivate at 37°C and 200rpm until the cell OD 600 =0.6~0.8, add inducer IPTG to a final concentration of 0.8mmol / L, induce culture at 28°C and 200rpm for 4~6h.
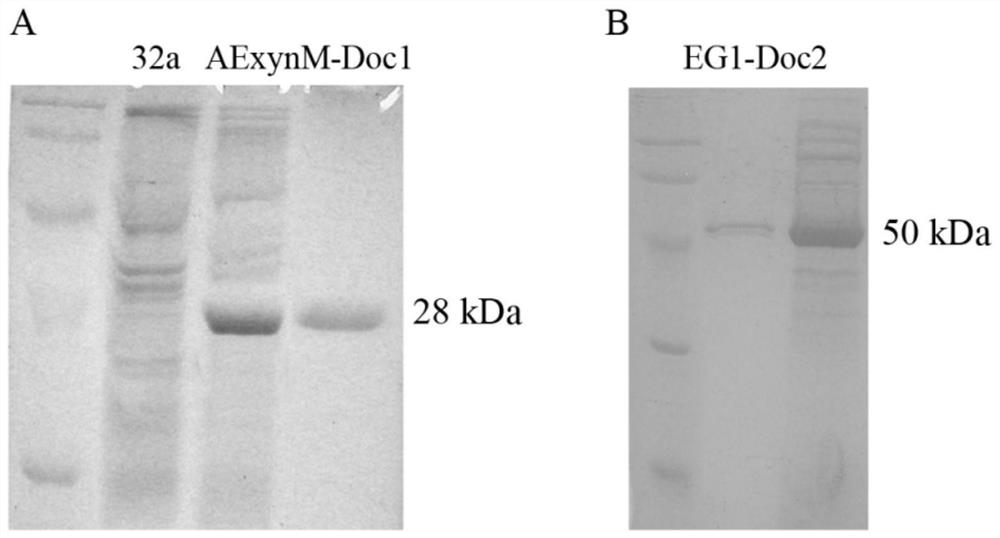
[0046] Take 30mL of the above induced culture, centrifuge at 8,000rpm for 10min, collect the bacteria, and use Na 2 HPO 4 - Citrate buffer (pH 5.5) containing 10mM CaCl 2 , wash the precipitate, and ultrasonically disrupt the cells in an ice bath, centrifuge at 15,000 rpm for 10 minutes, collect the supernatant, which is the crude enzyme solution, and perform enzyme activity determination and SDS-PAGE detection....
Embodiment 3
[0051] Cohesin display on the surface of Saccharomyces cerevisiae
[0052] 1) Construction of recombinant expression vector pYD1-Coh1-Coh2
[0053] The coding genes of cohesion proteins Coh1 and Coh2 are derived from R. flavefaciens (GenBank: AM262974) and R. flavefaciens (GenBank: 5M2O-A) respectively, and the specific sequences are obtained by artificial synthesis, and a connecting peptide is inserted between the two GGGGSGGGGSGGGGS, the nucleotide and amino acid sequences of Coh1-Coh2 are shown in SEQ ID NO.5 and SEQ ID NO.10, and this sequence is inserted between the EcoRI and XhoI sites of the surface display plasmid pYD1 to obtain pYD1-Coh1- Coh2.
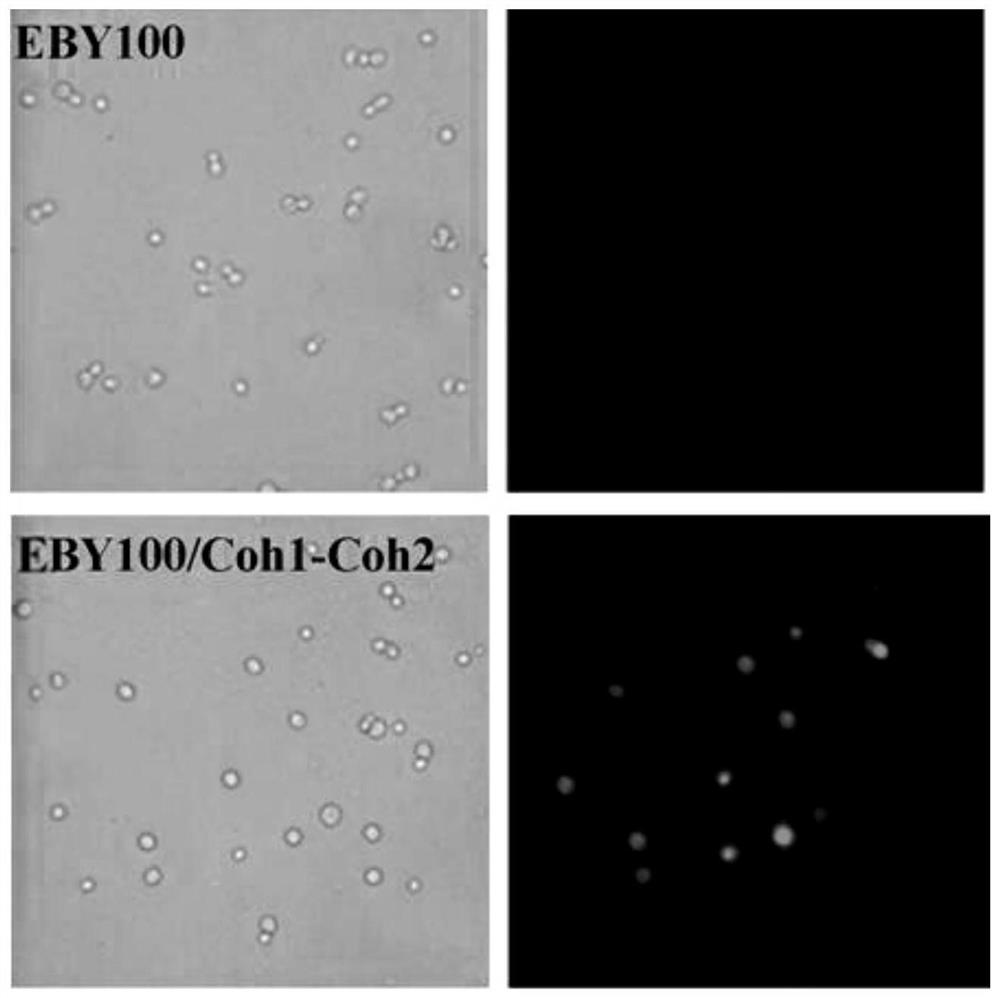
[0054] 2) Surface display of cohesin Coh1-Coh2
[0055] The lithium acetate chemical method was used to transform pYD1-Coh1-Coh2 into competent cells of Saccharomyces cerevisiaeEBY100, and the yeast recombinants were screened on a tryptophan selection plate, and the genomic DNA of the recombinants was extracted. Using this...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com