Escherichia coli engineering bacteria with improved acid stress resistance and application of escherichia coli engineering bacteria
A technology of Escherichia coli and acid stress, applied in the field of microbial engineering, can solve the problems of easy degradation of bacteria, accumulation of by-products, osmotic stress, etc., and achieve the effect of simple operation, improved acid stress resistance, and improved acid stress resistance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
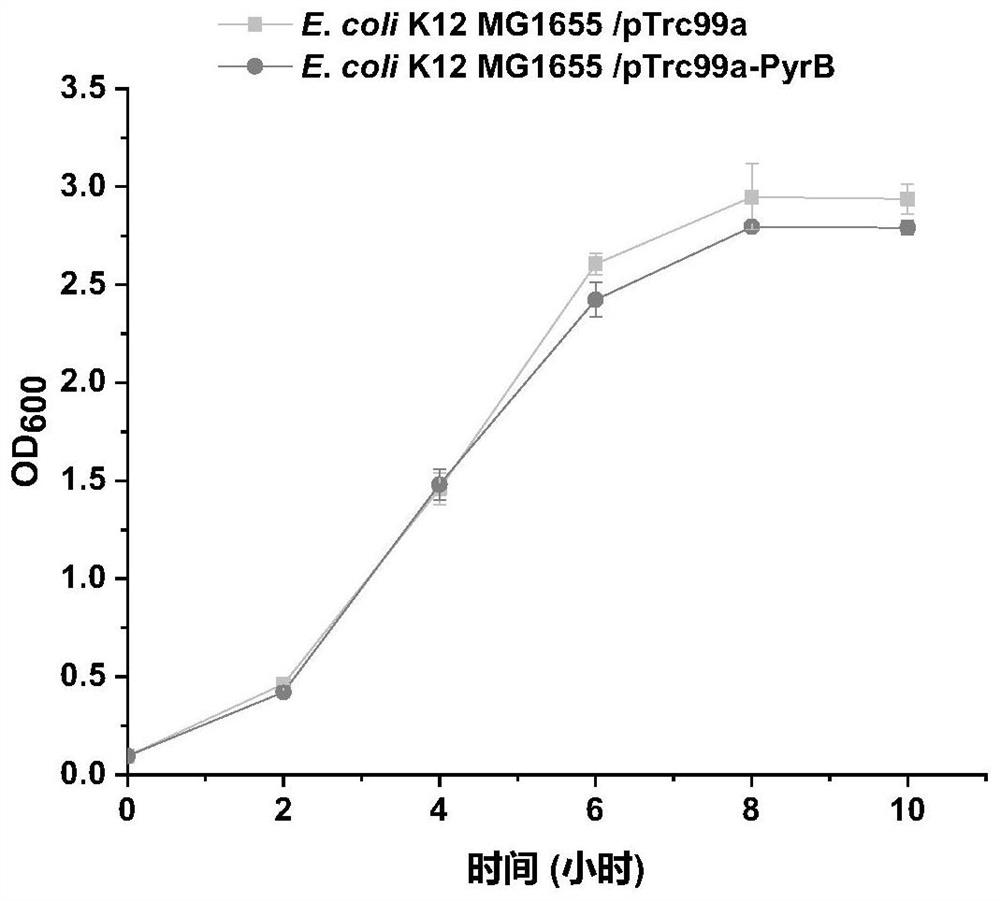
[0059] Example 1: Construction of recombinant strain E.coli K12 MG1655 / pTrc99a-PyrB
[0060] Specific steps are as follows:
[0061] (1) Based on the pyrB gene sequence in the NCBI database (encoding the catalytic subunit of aspartate carbamoyltransferase, the PyrB gene participates in the pyrimidine metabolism pathway and regulates aspartate carbamoyl transfer) designed as SEQ ID NO. 4 and the primer PTrc99a-PyrB-F shown in SEQ ID NO.5, PTrc99a-PyrB-R;
[0062] (2) Design the primers loop p-pTrc99a-F and loop p-pTrc99a-R shown in SEQ ID NO.10 and SEQ ID NO.11 respectively;
[0063] (3) Using the genome of E.coli K12 MG1655 as a template, using PTrc99a-PyrB-F and PTrc99a-PyrB-R as primers to amplify by PCR to obtain the gene fragment shown in SEQ ID NO.1;
[0064] (4) Using the vector pTrc99a as a template, using loop p-pTrc99a-F and loop p-pTrc99a-R as primers to amplify by PCR to obtain a linearized long fragment of the vector;
[0065] (5) Ligate the PCR products obtaine...
Embodiment 2
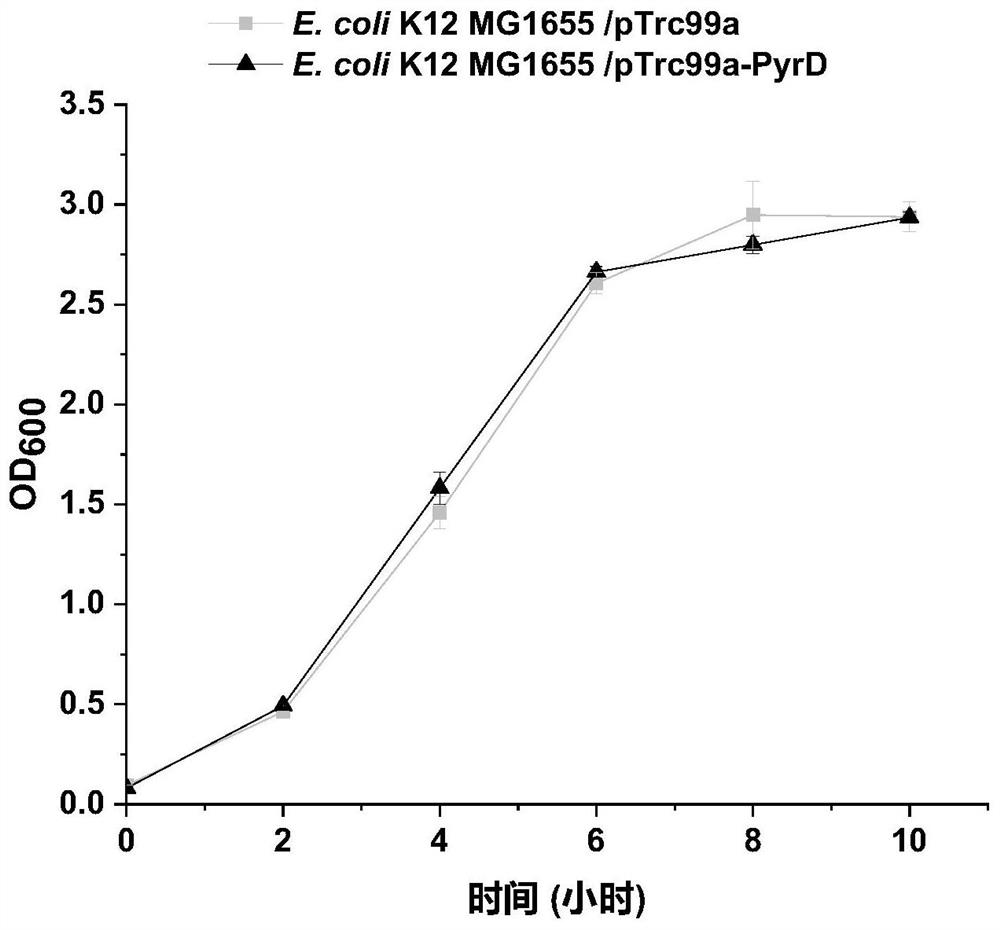
[0067] Example 2: Construction of recombinant strain E.coli K12 MG1655 / PTrc99a-PyrD
[0068] Specific steps are as follows:
[0069] (1) Based on the pyrD gene sequence in the NCBI database (the nucleotide sequence gene encoding dihydroorotate dehydrogenase PyrD, which participates in the pyrimidine metabolic pathway and regulates the dehydrogenation process of dihydroorotate), the design is as follows: SEQ ID Primers PTrc99a-PyrD-F and PTrc99a-PyrD-R shown in NO.6 and SEQID NO.7;
[0070] (2) Design the primers loop p-pTrc99a-F and loop p-pTrc99a-R shown in SEQ ID NO.10 and SEQ ID NO.11 respectively;
[0071] (3) Using the genome of E.coli K12 MG1655 as a template, using PTrc99a-PyrD-F and PTrc99a-PyrD-R as primers to amplify by PCR to obtain the gene fragment shown in SEQ ID NO.2;
[0072] (4) Using the vector pTrc99a as a template, using loop p-pTrc99a-F and loop p-pTrc99a-R as primers to amplify by PCR to obtain a linearized long fragment of the vector;
[0073](5) Liga...
Embodiment 3
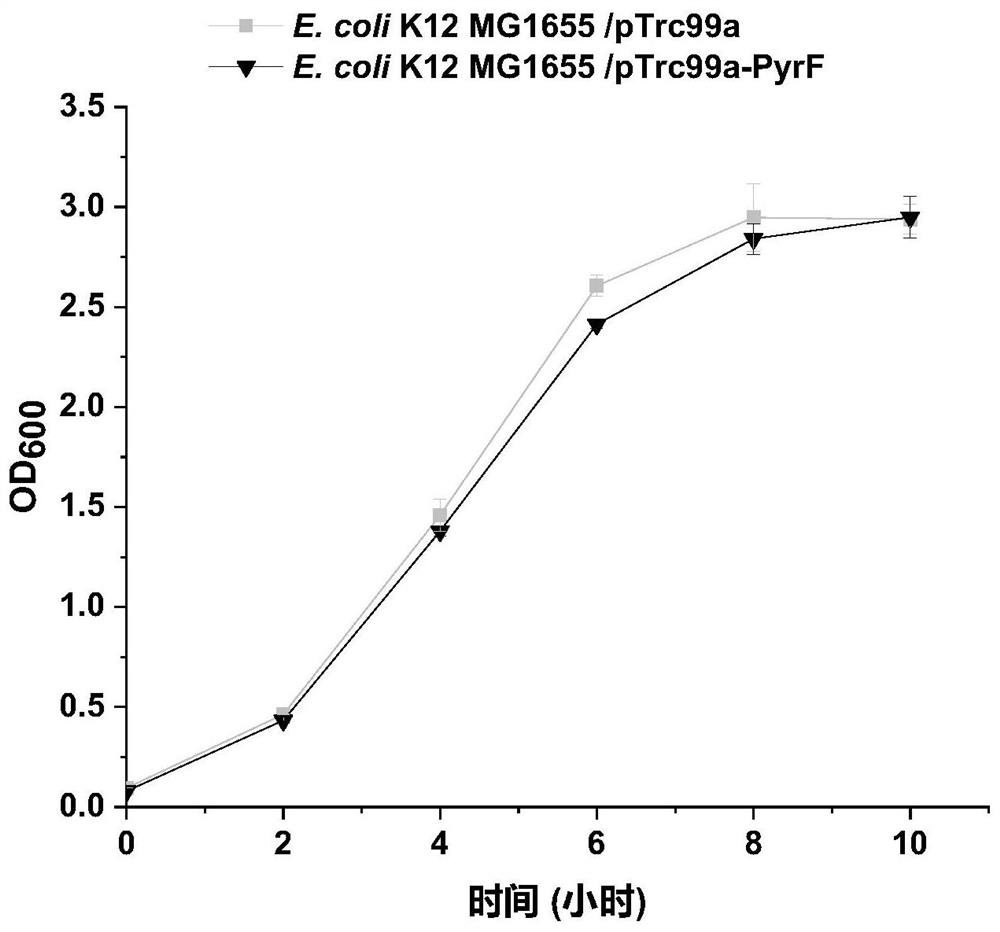
[0075] Example 3: Construction of recombinant strain E.coli K12 MG1655 / PTrc99a-PyrF
[0076] Specific steps are as follows:
[0077] (1) Based on the pyrF gene sequence in the NCBI database (encoding Orotidine-5'-phosphate decarboxylase protein PyrF gene, participates in pyrimidine metabolism pathway, regulates the decarboxylation of Orotidine-5'-phosphate) design such as SEQ ID NO.8 and Primers PTrc99a-PyrF-F and PTrc99a-PyrF-R shown in SEQ ID NO.9;
[0078] (2) Design the primers loop p-pTrc99a-F and loop p-pTrc99a-R shown in SEQ ID NO.10 and SEQ ID NO.11 respectively;
[0079] (3) Using the genome of E.coli K12 MG1655 as a template and using PTrc99a-PyrF-F and PTrc99a-PyrF-R as primers to obtain the gene fragment shown in SEQ ID NO.3 by PCR amplification;
[0080] (4) Using the vector pTrc99a as a template, using loop p-pTrc99a-F and loop p-pTrc99a-R as primers to obtain a linearized long fragment of the vector by PCR amplification;
[0081] (5) Ligate the PCR products o...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com