Target species specific sequence acquisition method and target species detection method
A technology of target species and acquisition method, applied in sequence analysis, bioinformatics, informatics, etc., can solve the problems of high requirements, large amount of calculation, long time, etc., and achieve high reliability, high accuracy, and simple method Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0038] Toxoplasma identification:
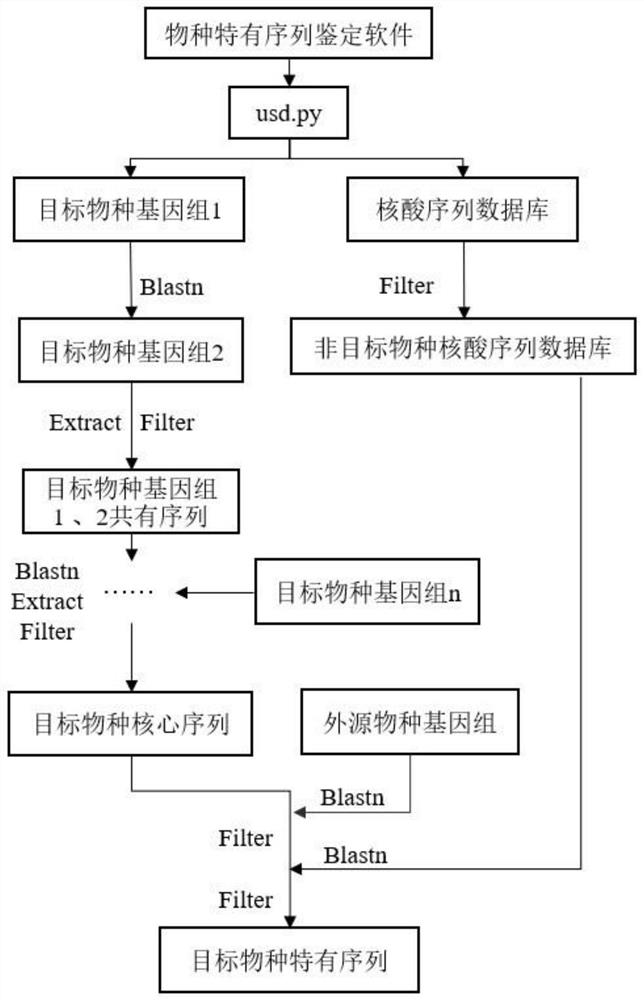
[0039] Taking GCA_000006565.2 (Toxoplasma gondii ME49) as the reference genome, plus other genomes of Toxoplasma gondii (toxoplasma classification id--5811, 5 genomes), together as the whole genome sequence of Toxoplasma gondii, and by analyzing the Toxoplasma gondii Alignment of all the whole genome sequences, screening nucleic acid sequences with a sequence similarity of 100%, to obtain the core sequence of Toxoplasma gondii;
[0040]The cat genome (version 9) was used as the genome database of exogenous species, and all nucleic acid sequences of Toxoplasma gondii in NCBI's NT database were filtered out according to the classification number of Toxoplasma gondii. (This step first excludes the influence of cats (toxoplasma hosts) individuals, filters out a part of the core sequence of toxoplasma gondii, and improves the subsequent comparison efficiency with the NT nucleic acid sequence database; in addition, if there are more genomes of for...
Embodiment 2
[0045] The method for obtaining the specific sequence of Brucella canis (Brucella Canis), the method for obtaining the specific sequence of Brucella canis comprises the following steps:
[0046] Select the whole genome sequence of Brucella canis in the NCBI genome database, and by comparing the whole genome sequence of said Brucella canis, screen out the core sequence of Brucella canis; All the whole genome sequences of Rutella are compared, and the nucleic acid sequences with a sequence similarity of 100% are screened to obtain the core sequence of Brucella canis.
[0047] Filter out all nucleic acid sequences of the described Brucella canis (taxid--36855) in the NT nucleic acid database to obtain the non-Brucellus canis nucleic acid sequence database;
[0048] Taking Brucella melitensis, Brucellasuis, Brucellaovis, and canine genomes as exogenous species, first compare the core sequence of Brucella canis with the genome of the exogenous species, and filter out the nucleic ac...
Embodiment 3
[0051] The method for obtaining the unique sequence of canine parvovirus (Canine parvovirus), the method for obtaining the unique sequence of canine parvovirus comprises the following steps:
[0052] Select the whole genome sequence of canine parvovirus in the NCBI genome database, and screen out the core sequence of canine parvovirus by comparing the whole genome sequence of the canine parvovirus; wherein, all the whole genome sequences of the canine parvovirus According to the alignment, the nucleic acid sequences with a sequence similarity of 100% are screened to obtain the canine parvovirus core sequence.
[0053] Filter out all nucleic acid sequences of the canine parvovirus (classification id--10788) in the NT nucleic acid database to obtain a non-canine parvovirus nucleic acid sequence database;
[0054] Taking Chiropteranprotoparvovirus, Eulipotyphlaprotoparvovirus, and canine genome as exogenous species, first compare the canine parvovirus core sequence with the exoge...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com