Method for improving PNA-based PCR inhibition efficiency, and application
A technology of inhibition efficiency and number of bases, applied in biochemical equipment and methods, microbiological measurement/inspection, etc., can solve the problems of small number of DNA fragments, large background DNA residue, and difficulty in detecting DNA fragments, so as to improve efficiency Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
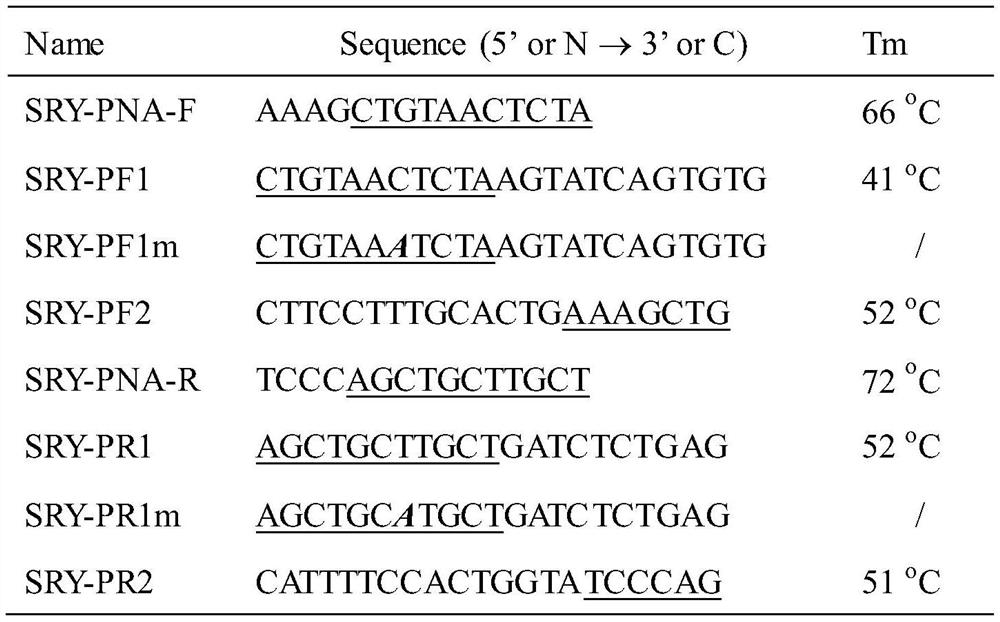
[0017] Design SRY-PF1 and SRY-PR1 primer sets, as shown in Table 1, the restriction site is between AAAG and CTGTAACTCTA, that is, before the first base of the overlapping sequence. According to the usual theory, attention should be paid to the PNA of the overlapping sequence The / DNA Tm value should be 3-5°C lower than the corresponding primer / DNA Tm value.
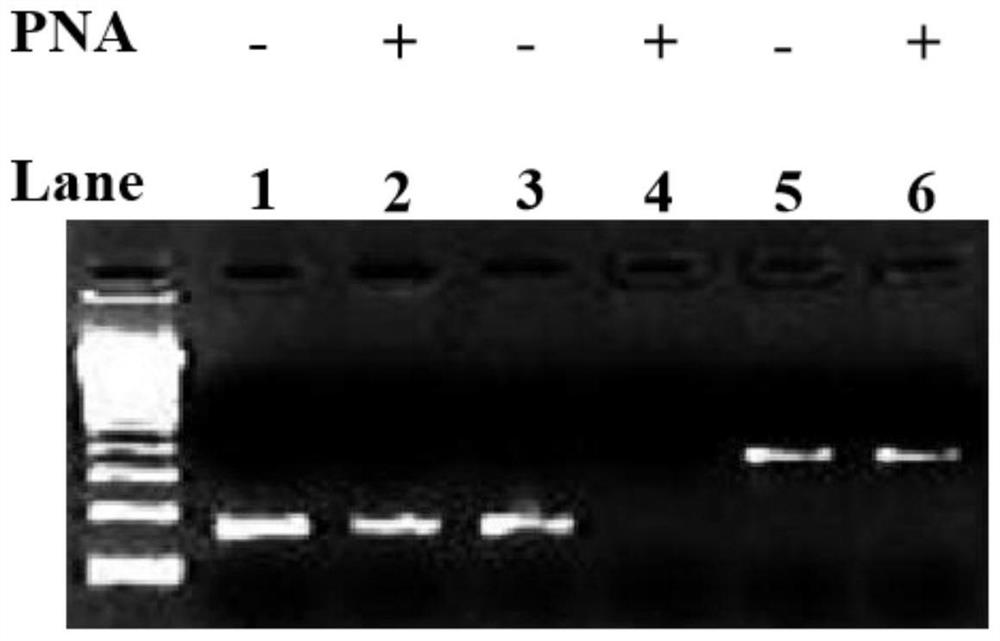
[0018] PCR reactions were performed in the absence (-) or presence (+) of the corresponding PNAs (SRY-PNA-F and SRY-PNA-R). Electrophoresis results such as figure 1 As shown, it can be seen that PNA cannot effectively inhibit PCR amplification (swimming lanes 1&2, figure 1 , the substrates of lanes 1-6 are all adult male whole blood DNA genomic DNA extracted by the general method, without enzyme digestion).
[0019] Table 1. Sequences of PNA and PCR primers used
[0020]
[0021] Single-base mismatch primers SRY-PF1m and SRY-PR1m were designed. The mismatch site was located in the middle of the overlapping sequence...
Embodiment 2
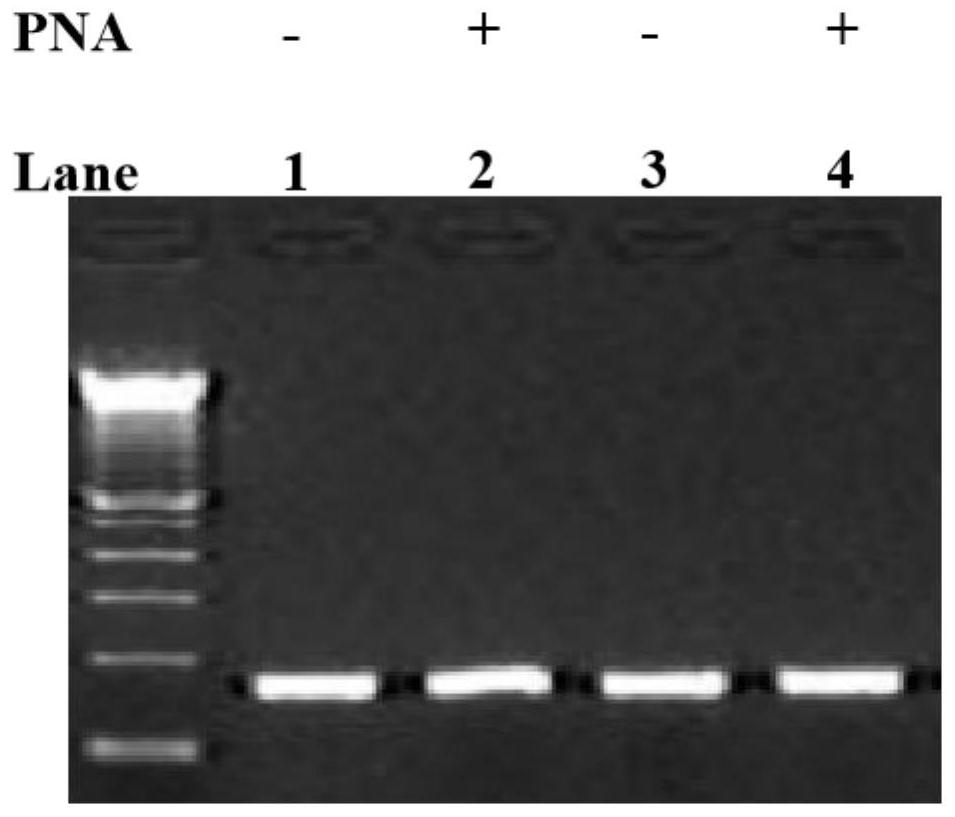
[0026] To generate short DNA fragments, the SRY gene was digested by restriction endonuclease Alu1. Enzymatic reaction: 100 units of Alu 1 digested 1 μg of genomic DNA at 37°C for 16 hours. The enzymatic reaction was terminated by heating to 65°C for 20 minutes. After the enzymatic reaction, in the absence (-) or presence (+) of PNA, different PCR primer sets were used to amplify the digested SRY gene, and the results were as follows figure 2 shown. SRY-PF1 and SRY-PR1 are perfectly complementary to the target sequence in the template, while SRY-PF1m and SRY-PR1m have a single base mismatch with the template. figure 2 It shows that single base mismatch primers do not affect the amplification of short DNA fragments. The short DNA here refers to the short DNA after digestion.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com