Prognostic marker and prognostic risk assessment model for metastatic colon adenocarcinoma and application of prognostic marker and prognostic risk assessment model
A risk assessment model and prognostic marker technology, applied in the field of genetic technology and medicine, can solve the problem of not providing differential genes, and achieve the effect of good discrimination performance
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0069] The purpose of this example is to screen DEGs in patients with primary COAD and metastatic COAD.
[0070] (1) Screening DEmiRNAs and their target genes
[0071] In order to screen DEmiRNAs from the miRNA-Seq dataset of the TCGA database, differential expression analysis was performed using the Edger software package in this example.
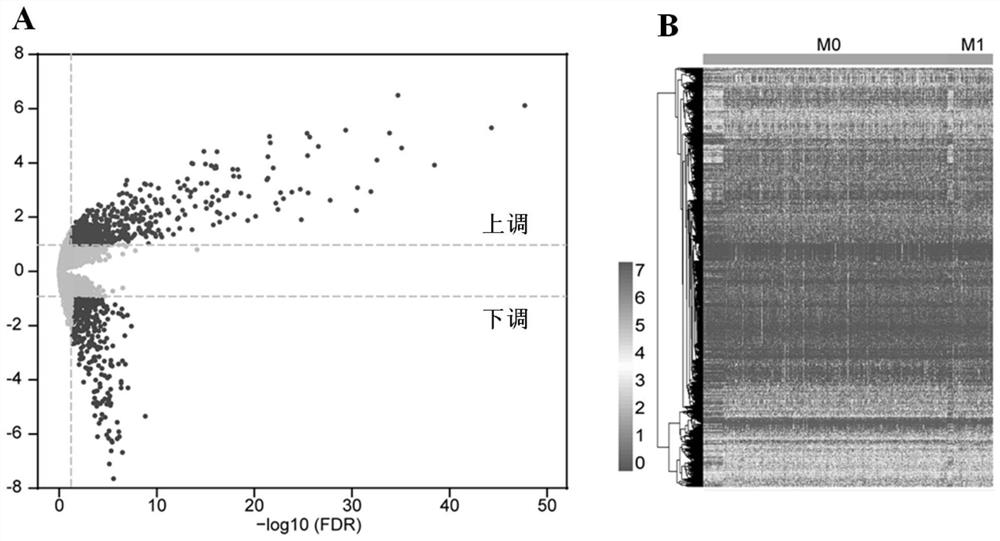
[0072] In this database, 62 / 379 (16.4%) of DEmiRNAs came from patients with M1 stage, and 317 / 379 (83.6%) came from patients with M0 stage, where M0 means patients without distant metastasis, and M1 means patients with distant metastasis. The 14 DEmiRNAs with critical points |log2FC|>1 and p-valuefigure 1 As shown, as shown in Figure A, there are 8 up-regulated DEmiRNAs and 6 down-regulated DEmiRNAs, and the corresponding heat map, that is, the specific miRNAs are shown in Figure B;
[0073] The specific analysis is shown in Table 1 below.
[0074] Table 1
[0075]
[0076]
[0077] Meanwhile, 8918 potential target genes of 8 up-r...
Embodiment 2
[0086] The purpose of this example is to screen genes related to the prognosis of metastatic COAD.
[0087] (1) Functional annotation and enrichment analysis
[0088] In order to analyze the enrichment pathway of DEmiRNAs, KEGG pathway enrichment analysis was carried out using DIANA bioinformatics tool in this example. These 14 DEmiRNAs were significantly enriched in cancer- and metastasis-related pathways, such as cell cycle, adherens junctions, and p53 signaling pathways.
[0089] In addition, pathway and process enrichment analyzes were performed on the basis of 127 candidate DEGs.
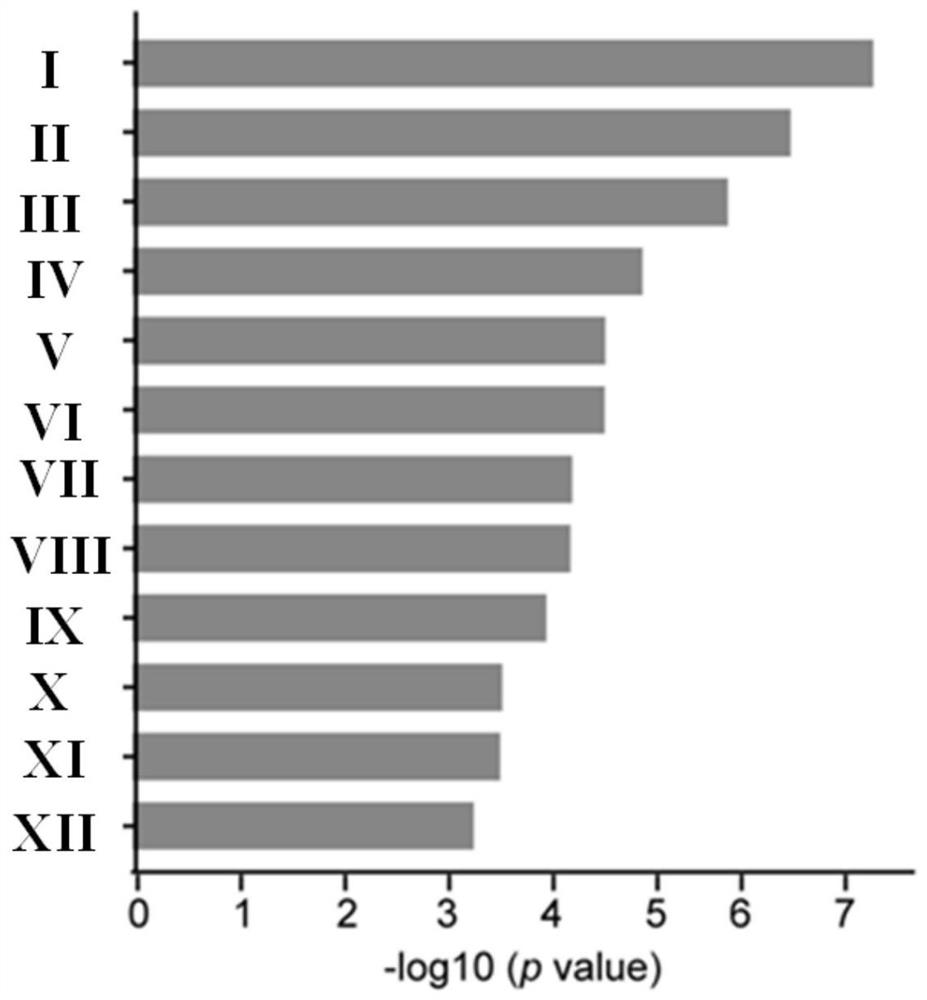
[0090] The first 12 involved clusters such as image 3 As shown, they are:
[0091] I. NABA matrisome associated (NABA matrisome associated); II. Forebrain neuron differentiation; III. Cell-cell adhesion through plasma membrane adhesion molecules (Cell-cell adhesion); IV. Skin development (skin development); V, blood circulation; VI, peptide cross-linking; VII, nervous system; VIII, forelim...
Embodiment 3
[0112] This example uses prognostic markers for risk scoring and survival analysis, and compares the performance of individual genes versus combined gene signature panels.
[0113] (1) A gene signature panel with prognostic value for COAD was established using stepwise multiple Cox regression analysis. According to the results, the risk scoring system for each patient was calculated as follows:
[0114] Risk score = LEP × 0.134582 + REG3A × (-0.05402) + DLX2 × 0.211469;
[0115] Patients were then classified into low-risk (n178) groups according to the median risk score.
[0116] Such as Figure 4 As shown, compared with the low-risk group, the survival period of patients in the high-risk group was significantly worse (log-rank test, p value 0.01692, p value <0.05).
[0117] In order to evaluate the prognostic value of the risk score, the ROC curve of OS at 5 years was drawn and the AUC value was calculated in this embodiment.
[0118] From Figure 5 It can be seen from th...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com