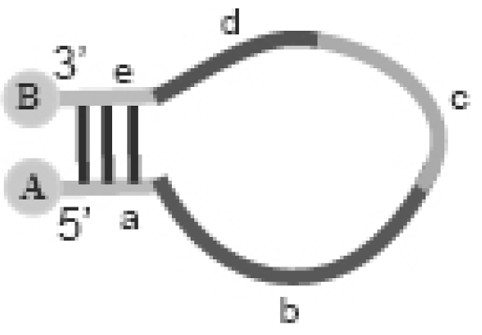
Modified fluorescent probe and application thereof
A technology of probes and oligonucleotide probes, applied in the field of modified fluorescent probes, which can solve the problems of low detection specificity and accuracy, difficult PCR, and many DNA damages and breaks
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
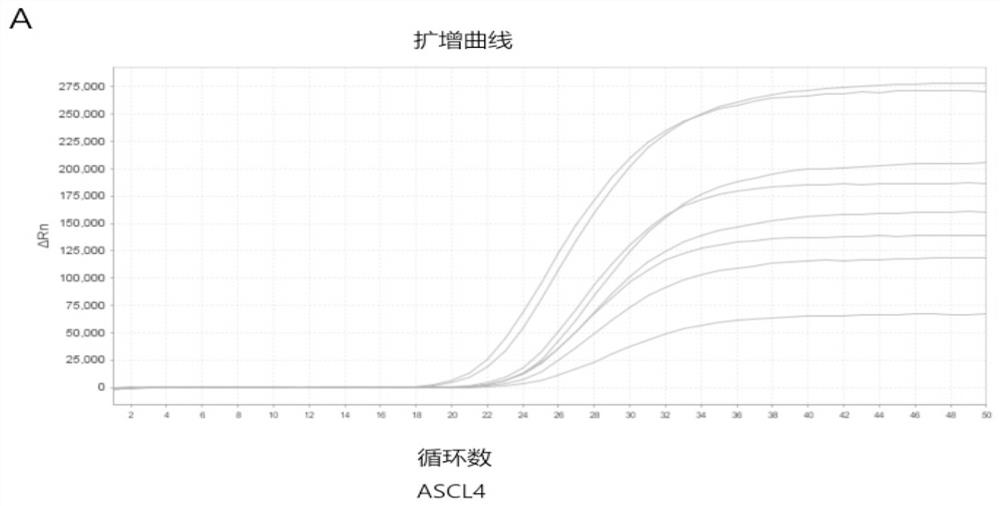
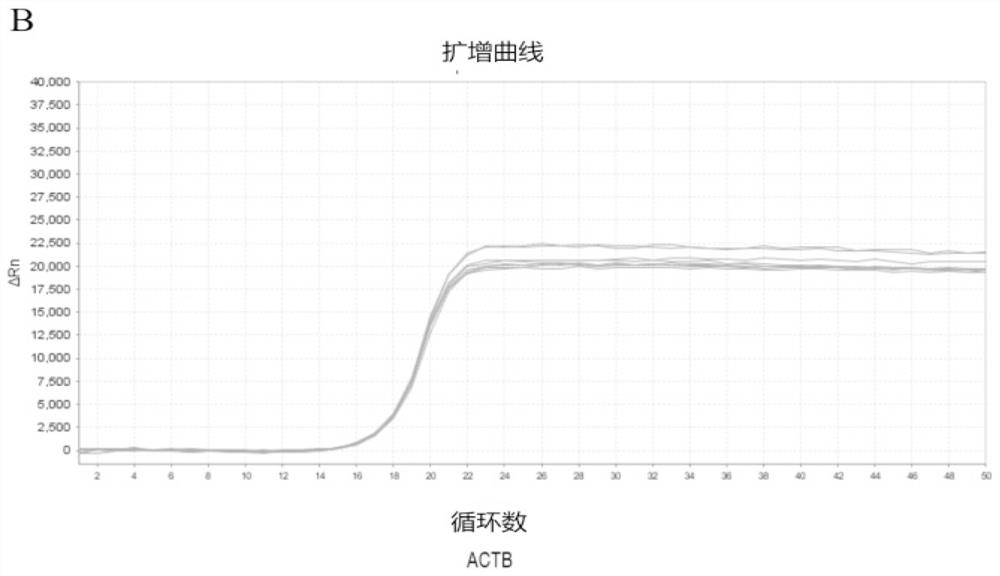
[0097] Example 1: The present invention designs probes for methylation-specific detection
[0098] Use bisulfite-treated reference DNA to verify primer specificity. First, in this example, the target DNA in the ASCL4 gene region was selected as the detection site, and the internal reference gene ACTB was designed, and primers and probes were designed respectively. Then, the gradient was configured with 100%, 50%, 25%, and 10% ratios of methylated DNA (Qiagen 59655) to unmethylated DNA (Qiagen 59665), and the total amount of DNA was 4 ng. Finally, the designed primers were used to amplify the DNA in each gradient DNA mixture in two replicates. Refer to Table 1 for the sequences of the primers and the modified probes of the present invention used to detect ASCL4, as well as the unmethylated specific primers and probes used to detect the internal reference gene ACTB. Among them, ASCL4 uses the invention to design methylation-specific probes, and the internal reference gene uses...
Embodiment 2
[0107] Embodiment 2: The design probe of the present invention compares with conventional Taqman probe
[0108] Compared with the detection method in Example 1, the detection performances of the designed probes of the present invention and conventional probes were compared. Wherein the routine probe sequence of ASCL4 is: TCGGCGTTTTCGTTTAGTAGC.
[0109] The gradient configuration was 100%, 50%, 25%, 10% ratio of methylated DNA (Qiagen 59655) to unmethylated DNA (Qiagen 59665), and the total amount of DNA was 4 ng. The designed primers were used to amplify the DNA of each gradient DNA mixture, which was repeated twice.
[0110] result
[0111] The results show that under the same template gradient, the probe of the present invention produces a better amplification curve than the conventional probe, showing a lower fluorescence background ( Figure 3A-3D ), higher fluorescence increments and smaller Ct values ( Figures 4A-4D ), and can significantly improve the amplificati...
Embodiment 3
[0114] Embodiment 3: The present invention design probe is used for the detection of colorectal tissue sample
[0115] Compared with the test method in Example 1, using the ASCL4 probe sequence designed by the present invention, and the internal reference detection primers and probe sequences in Example 1, 10 white blood cell DNA, paracancerous tissue DNA, and intestinal adenoma tissue were detected. DNA and colorectal cancer tissue DNA.
[0116] Described detection comprises the steps:
[0117] Ten DNA samples each were obtained from blood cells, paracancerous tissue, high-grade adenoma tissue, and colorectal cancer tissue (ie, a total of 40 samples). The commercial kit QIAamp DNA Mini Kit was used to extract blood cell DNA according to the instructions; for paracancerous tissue, high-grade adenoma tissue and colorectal cancer tissue DNA, the commercial kit QIAamp DNA FFPETissue Kit was used to extract according to the instructions;
[0118] Use the commercial bisulfite conve...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com