Cynoglossus semilaevis disease-resistant breeding gene chip and application thereof
A semi-smooth tongue sole and gene chip technology, applied in genomics, DNA/RNA fragments, instruments, etc., can solve the problem of lack of gene chips in the selection and breeding of semi-smooth tongue sole, and achieve the effects of improving the survival rate of offspring, efficient technical means, and speeding up the breeding process
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
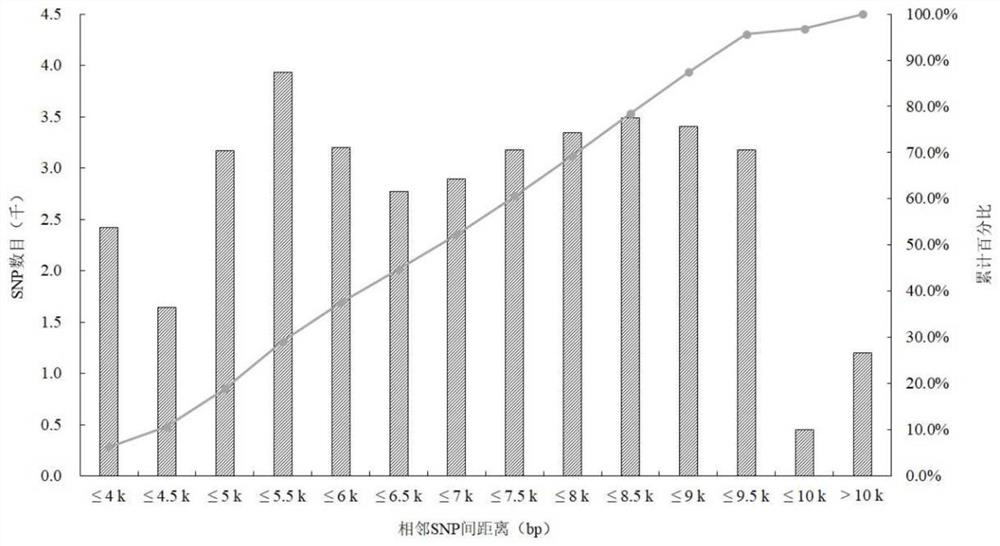
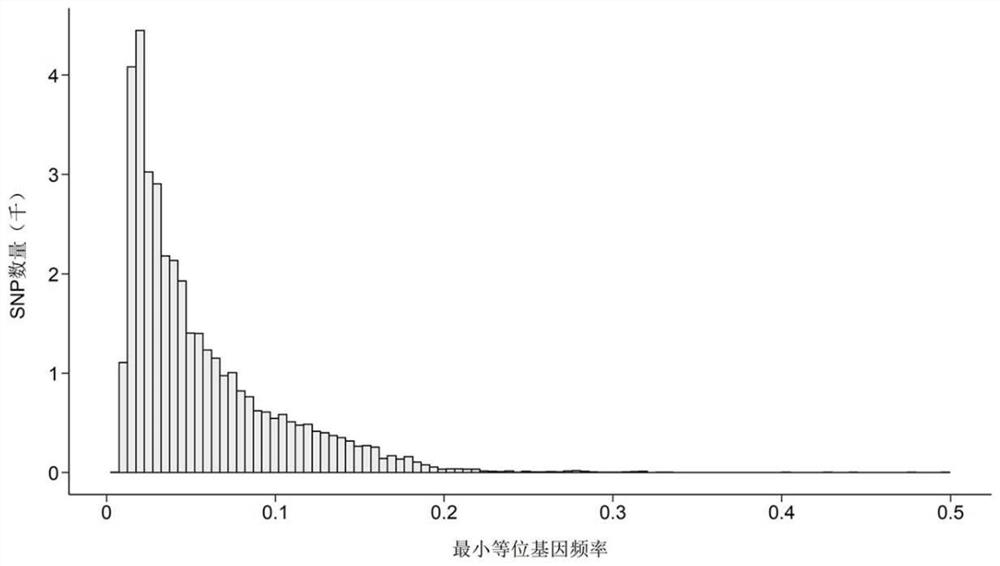
[0035] Example 1: SNP site screening and chip preparation of "Sole Core No. 1" gene chip
[0036] 1. Establishment of reference population and collection of phenotypic data of tongue sole resistant to Vibriosis harvei
[0037] The reference populations of tongue sole resistant to vibriosis harvei are all derived from the family of tongue sole selected for many years by the inventor's research group. The epidemiological investigation found that Vibrio harveyi is the main pathogenic bacteria in tongue sole aquaculture industry, and infected individuals will have symptoms such as rotten skin and rotten tail. In order to improve the ability of semi-smooth tongue sole to resist Vibrio harvei disease, our research group began to carry out the selection and breeding of semi-smooth tongue sole against Vibrio harvei disease through the artificial infection experiment of Vibrio harvei in 2012.
[0038] In 2014, 2016 and 2018, 11,207 tails of 86 families, 3,714 tails of 23 families and ...
Embodiment 2
[0052] Example 2: Evaluation of the Predictive Accuracy of Genomic Selection Methods Using Reference Populations and Microarray Design Loci
[0053] From the high-quality SNPs obtained in Example 1, extract the same SNPs as the chip design sites, use these SNPs to construct the genomic kinship matrix G matrix, and use 5-fold cross-validation to evaluate the prediction accuracy of the GBLUP method. In addition, the predictive accuracy of the BLUP method was also evaluated for comparison with that of the GBLUP method.
[0054] The analysis results showed that the prediction accuracy of GBLUP (0.780) was higher than that of the BLUP method (0.599) (Table 2, Figure 8 ). Therefore, the SNP loci designed by the "Sole Core 1" gene chip can meet the computational needs of genome selection, and can ensure the smooth development of genome selection breeding.
[0055] The specific operation method is as follows (Linux environment):
[0056] 1. Use PLINK software to extract the chip d...
Embodiment 3
[0072] Example 3: Method for using the "Sole Core No. 1" gene chip prepared in Example 1
[0073] Randomly select 24 resequenced fin ray samples of half-smooth tongue sole, and type these samples according to the following steps:
[0074] 1. Preparation of samples to be tested
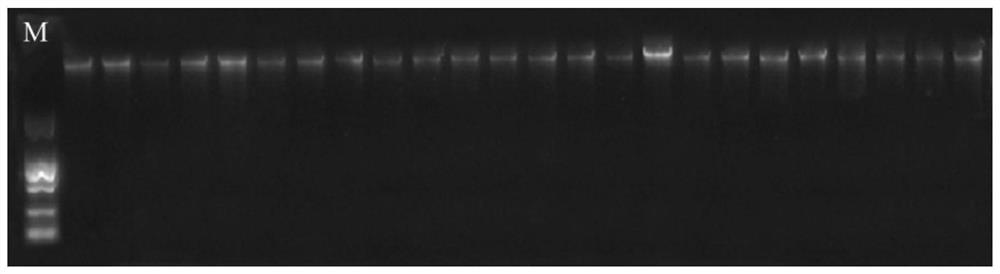
[0075] About 1 cm of caudal fin rays of half-smooth tongue sole were cut, and the genomic DNA of the test sample was extracted and purified using a DNA extraction kit (Tiangen, Beijing), and then the DNA quality and concentration were detected by 1% agarose gel electrophoresis and ultraviolet spectrophotometer. Observe under the gel imager after electrophoresis, if a single band appears in the DNA, the fragment length is greater than 10kb, the integrity is good, and there is no degradation, the quality of the genomic DNA is considered to be good ( image 3 ). In addition, using a spectrophotometer to detect, if the sample A260 / 280 is between 1.8 and 2.0, A260 / 230 is greater than 1.5, the concentratio...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com