Primer, probe and reagent kit for universal influenza A virus nucleic acid detection
A type of influenza A virus and kit technology, applied in biochemical equipment and methods, microorganisms, methods based on microorganisms, etc., can solve problems such as the difficulty in designing primers and probes, and achieve high detection rate, high sensitivity, and simple combined effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0016] Embodiment 1: Design and synthesis of primers and probes
[0017] One of the keys of the present invention is to design a set of primers and probes that can be used for general nucleic acid detection of influenza A virus. According to the genome sequence of influenza A virus in the GenBank database, use MAFFT software to compare and analyze the consistency of different types of gene sequences, select the relatively conserved regions of the sequence mutation sites of influenza A virus matrix protein M1 and M2 to design genome-wide specificity Amplification primer sequence,
[0018] The designed primers and probe sequences are:
[0019] The nucleotide sequence of the forward primer IFN-UN-F is shown in SEQ ID No: 1, namely: 5'-CAAGACCAATCCTGTCACCTYTR -3'; the nucleotide sequence of the reverse primer IFN-UN-R is shown in SEQ ID No: 2, namely: 5'-TCTACGCTGCAGTCCTCGCTC-3'; and the nucleotide sequence of the probe IFN-UN-P as shown in SEQ ID No: 3, namely: 5'-ACGCTCACCGTGC...
Embodiment 2
[0020] Embodiment 2: the extraction of RNA
[0021] Influenza A virus was cloned and sequenced in accordance with the "Molecular Cloning Experiment Guide (Third Edition)". The throat swabs, sputum, alveolar lavage fluid, tissue exudate, feces and urine collected from 60 patients with cold symptoms were used as samples to extract influenza virus cell cultures and use high-purity viral RNA reagents High Pure Viral RNA Kit (High Pure Viral RNA Kit) was used to extract RNA from influenza virus samples, and the specific extraction steps were performed according to the operating instructions. Among them, sputum, throat swab liquid and feces samples need to be pretreated, and 0.01M PBS, pH7.5, should be added to sputum and throat swab samples in advance; The suspension was centrifuged at 12000×g for 10 min, and the supernatant was taken for RNA extraction, eluted with 50 μl DEPC water, and stored at -80°C.
Embodiment 3
[0022] Embodiment 3: the establishment of Real-time RT-PCR amplification method
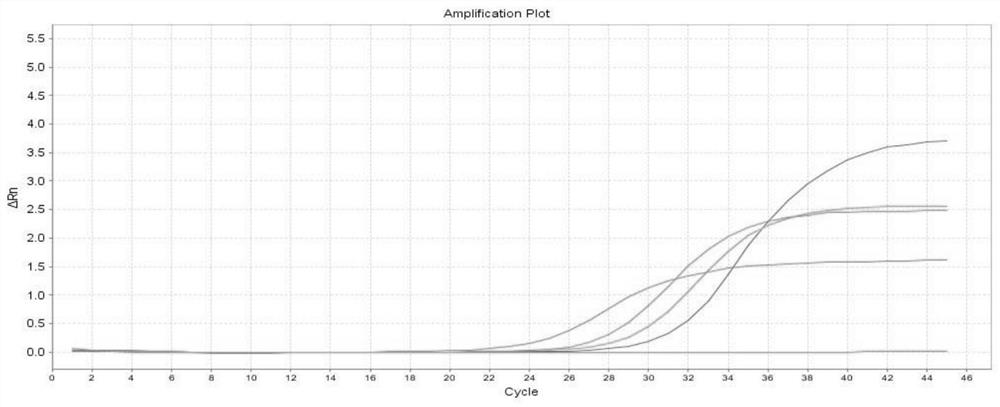
[0023] Using the influenza virus RNA extracted from each sample in Example 2 as a template, a real-time RT-PCR reaction was performed (reagents for the reaction were purchased from TaKaRa).
[0024] The fluorescent quantitative RT-PCR reaction system is as follows:
[0025] The 25μl rRT-PCR detection system contains: 2×OneStep RT-PCR Buffer 12.5μl, 10μM forward primer (SEQ ID No:1) 1.5μl, 10μM reverse primer (SEQ ID No:2) 1.5μl, 10μM probe Needle (SEQ ID No:3) 0.5μl, Rox Reference Dye Ⅱ (50×) 0.5μl, 5U / μl Ex Taq HS 0.5μl, 40U / μl PrimeScriptRT enzyme Mix 0.5μl, sample RNA to be tested 5μl, RNase Free dH 2 O 2.5 μl. Put the reaction tube of the above reaction system into a fluorescent quantitative PCR instrument, and set the rRT-PCR reaction conditions as follows: reverse transcription at 50°C for 20 minutes, reaction at 95°C for 3 minutes, denaturation at 95°C for 15 seconds, annealing / extension...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com