Nucleic acid sequence combination, kit and detection method for LAMP-CRISPR isothermal detection of prawn enterocytozoon hepatopenaei
A technology of LAMP-CRISPR and hepatic enterocystosis, applied in the field of biotechnology detection, can solve the problem of low sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
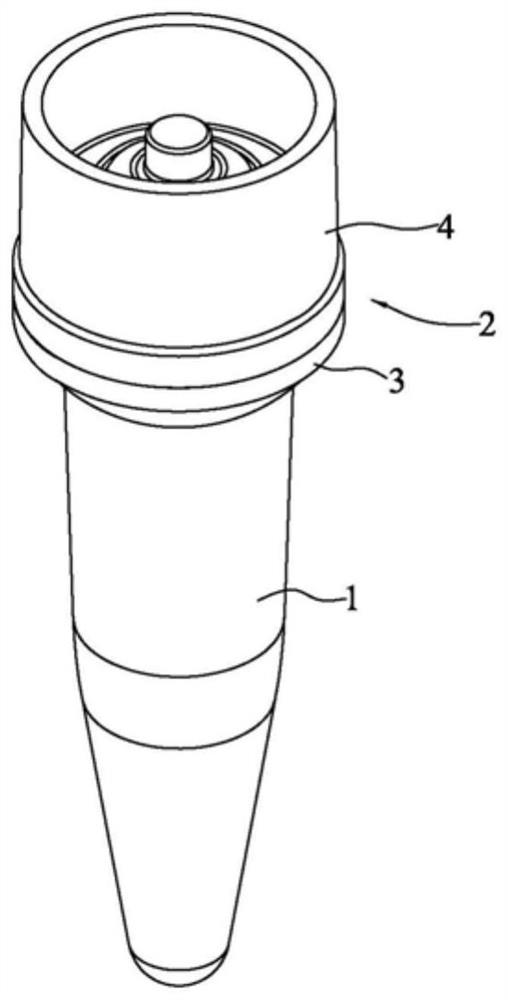
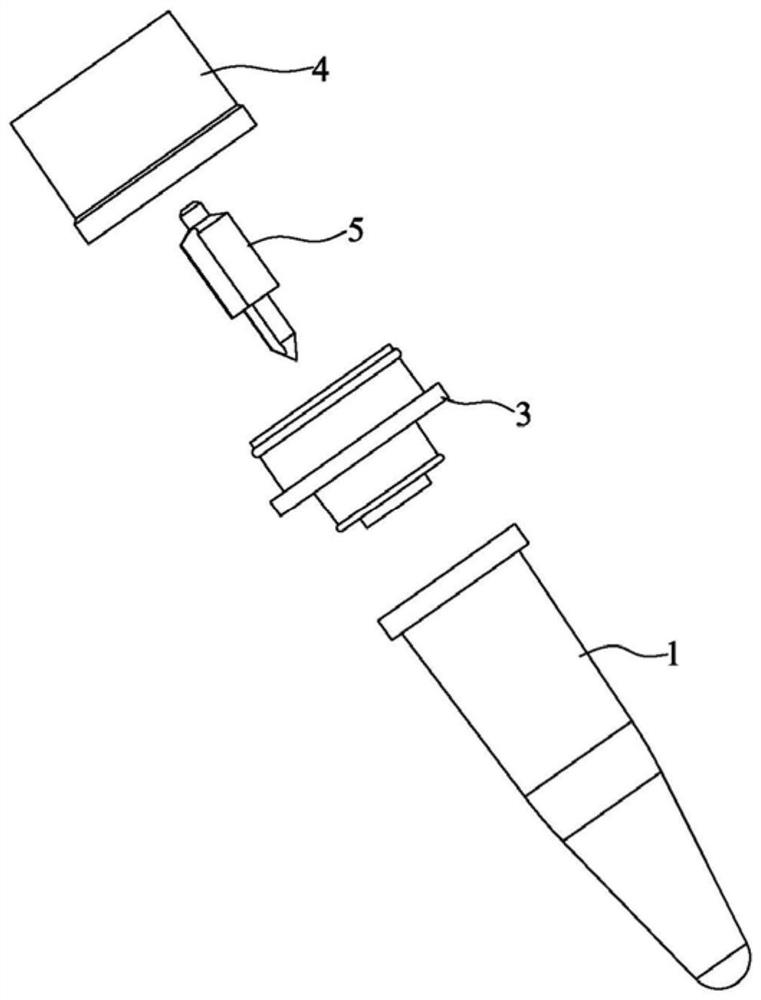
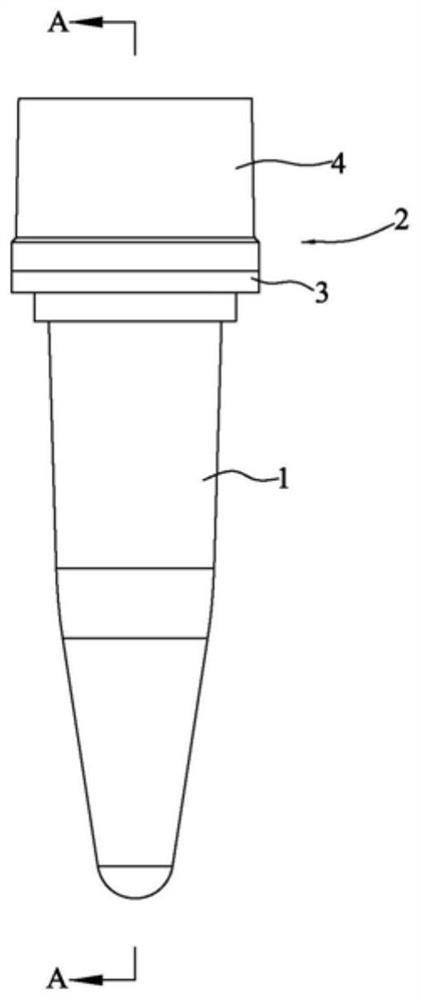
[0069] like Figure 1-13 As shown, a reaction tube for nucleic acid amplification and detection in a two-step method includes a tube body 1 for adding the first reaction solution and a cover body 2 that cooperates with the tube body 1 to seal. A liquid box 3 for temporarily storing the second reaction solution during a one-step reaction. The bottom surface of the liquid box 3 has a first opening 31 for the second reaction solution to flow into the tube body 1. The top surface of the liquid box 3 is provided with The second opening 32 for adding the second reaction solution, and the first opening 31 is sealed with a sealing film before adding the second reaction solution. The overall structure of the tube body 1 is similar to that of a conventional PCR reaction tube, and the bottom is used for adding reaction solution for reaction.
[0070] like Figure 5 and 7 As shown, the liquid box 3 has a plug portion 33 extending into the tube body 1 and plugged with the mouth of the t...
Embodiment 2
[0080] 1. Experimental materials
[0081] Actual sample 1: No. ND01, sampled from shrimp seedlings in the Ningde shrimp seedling breeding base in 2019;
[0082] Actual sample 2: No. HZ02, sampled in 2019 from juvenile shrimps in Hangzhou Shrimp Breeding Base;
[0083] Actual sample 3: No. HZ03, sampled in 2019 from the juvenile shrimp in the shrimp seedling breeding base in Hangzhou;
[0084] Actual sample 4: No. FZ04, sampled from juvenile shrimp in Fuzhou Shrimp Breeding Base in 2019;
[0085] Actual sample 5: No. FZ05, juvenile shrimp sampled at the Fuzhou Shrimp Breeding Base in 2019;
[0086] Actual sample 6: No. FZ06, sampled from juvenile shrimp in the Fuzhou Shrimp Breeding Base in 2019.
[0087] 2. Extraction of Shrimp Iridovirus DNA
[0088] (1) Weigh 30 mg of experimental samples (parotid glands for sample 1, whole shrimp seedlings for sample 2), add 500 μL of physiological saline, remove the liquid after mixing upside down, and repeat this step once;
[0089] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com