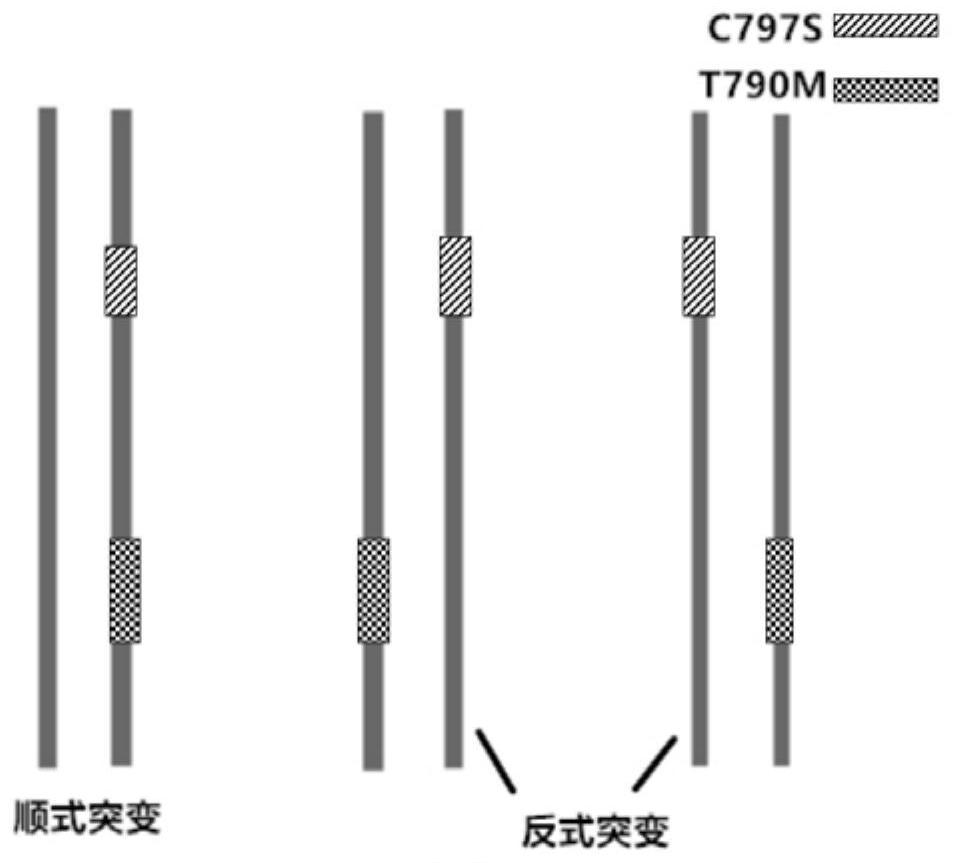
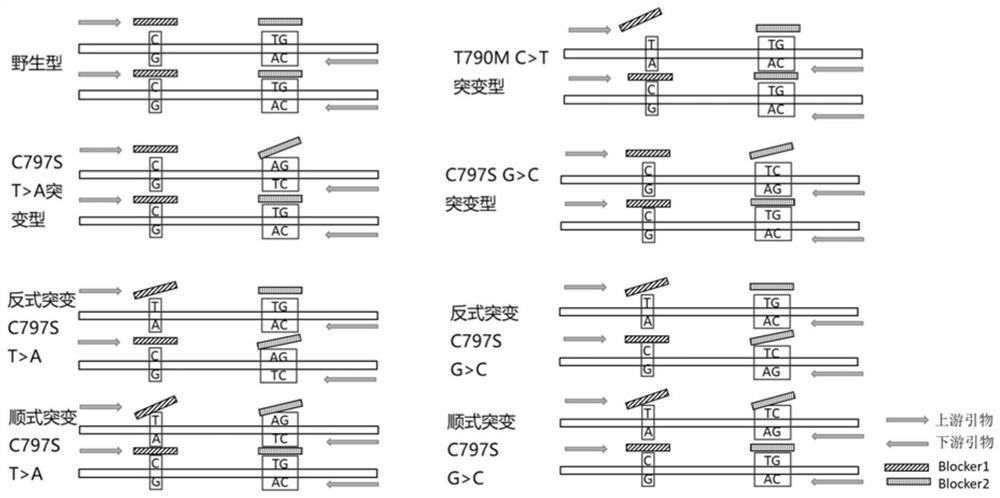
Methods and probes for detecting the cis-trans mutant configuration of egfr-t790m and egfr-c797s
A technology of EGFR-T790M and EGFR-C797S, which is applied in biochemical equipment and methods, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problems of high detection sensitivity, inability to directly judge the configuration, and high cost. Sensitive effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0139] This example provides a kit for detecting EGFR-T790M and EGFR-C797S cis-trans mutation types, the composition of which is shown in the following table:
[0140] Table 2
[0141]
[0142]
[0143] (2) The experimental procedure of this kit is as follows
[0144] table 3
[0145]
[0146] (1) Comparison of Blocker 1 blocking performance:
[0147] Plasmid DNA with different mutation information was used as template, and the plasmid DNA information is shown in the following table:
[0148] Table 4
[0149] name type of mutation Plasmid 001 Wild type (WT) Plasmid 002 T790M C>T Plasmid 003 C797S T>A Plasmid 004 C797S G>C Plasmid 005 T790M C>T&C797S T>A Plasmid 006 T790M C>T&C797S G>C
[0150] The system was prepared according to the formula in the following table, and the PCR program was shown in Table 3.
[0151] table 5
[0152]
[0153]
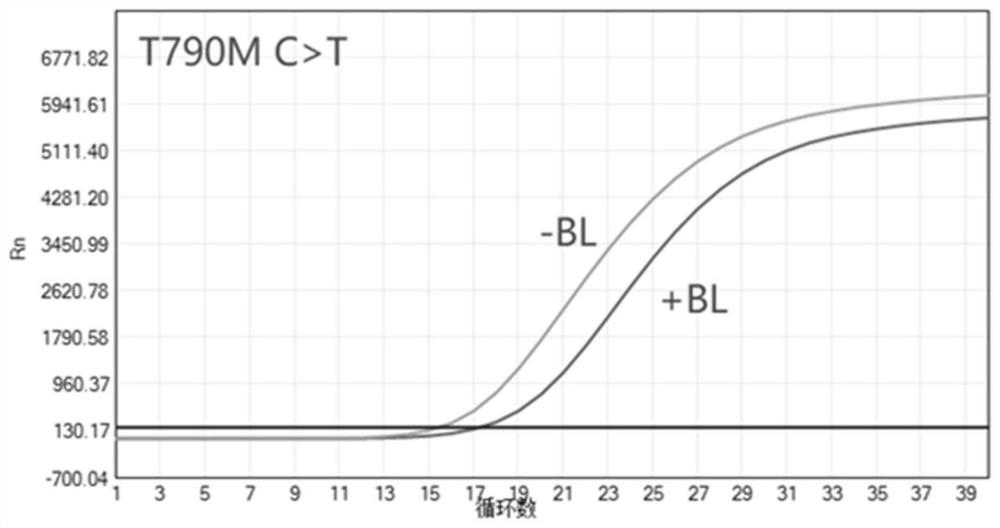
[0154] The result is as Figures 3 to 8 As shown, Blocke...
Embodiment 2
[0196] This embodiment provides a kit for detecting cis-trans mutation types of EGFR-T790M and EGFR-C797S, which differs from the embodiment in that the following probes are used:
[0197] Table 11
[0198]
[0199] Plasmid DNA with different mutation information was used as template, and the plasmid DNA information is shown in the following table:
[0200] Table 12
[0201] name type of mutation Plasmid 001 Wild type (WT) Plasmid 002 T790M C>T Plasmid 003 C797S T>A Plasmid 004 C797S G>C
[0202] The system was prepared according to the formula in the following table, and the PCR program was shown in Table 3.
[0203] Table 13
[0204]
[0205] The product was sent for inspection and first-generation sequencing was performed: the results are as follows
[0206] Plasmid 001 (wild type) Blocker 3 product sequencing results are as follows Figure 55 shown; Blocker 4 product sequencing results are as follows Figure 56 Shown: B...
Embodiment 3
[0212] This embodiment provides a kit for detecting cis-trans mutation types of EGFR-T790M and EGFR-C797S, which differs from the embodiment in that the following probes are used:
[0213] Table 14
[0214]
[0215] The system was prepared according to the formula in the following table, the DNA samples were shown in Table 4, and the PCR program was shown in Table 3.
[0216] Table 15
[0217]
[0218] Blocker5 amplification results are as follows Figures 63 to 68 Shown: Blocker 5 blocked wild-type and single-site C797S mutant DNA amplification, but not or less blocked single-site T790M mutation and cis-mutation T790M C>T&C797S T>A or G>C DNA amplification.
[0219] Blocker6 amplification results are as follows Figures 69 to 74 As shown, Blocker 6 blocked wild-type and single-site T790M mutant DNA amplification, and did not block or less blocked single-site C797S T>A or G>C and cis-mutated T790M C>T&C797S T>A or G>C DNA amplification.
[0220] Comparison of the bl...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com