A molecular modification design method for improving the catalytic efficiency of prolyl endopeptidase
A technology of prolyl endopeptidase and catalytic efficiency, which is used in the analysis of two-dimensional or three-dimensional molecular structure, sequence analysis, instruments, etc., can solve the problems of poor pH stability and inapplicability, and achieve the effect of improving catalytic activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032]Example 1: Homology modeling and molecular docking of Sphaerobacter thermophiles prolyl endopeptidase
[0033] 1. Arrange and compare the enzymatic properties of prolyl endopeptidases of different species, select the prolyl endopeptidase derived from Sphaerobacter thermophiles as the object of transformation, the amino acid sequence of the prolyl endopeptidase As shown in SEQ ID NO.1, its nucleotide sequence is shown in SEQ ID NO.2.
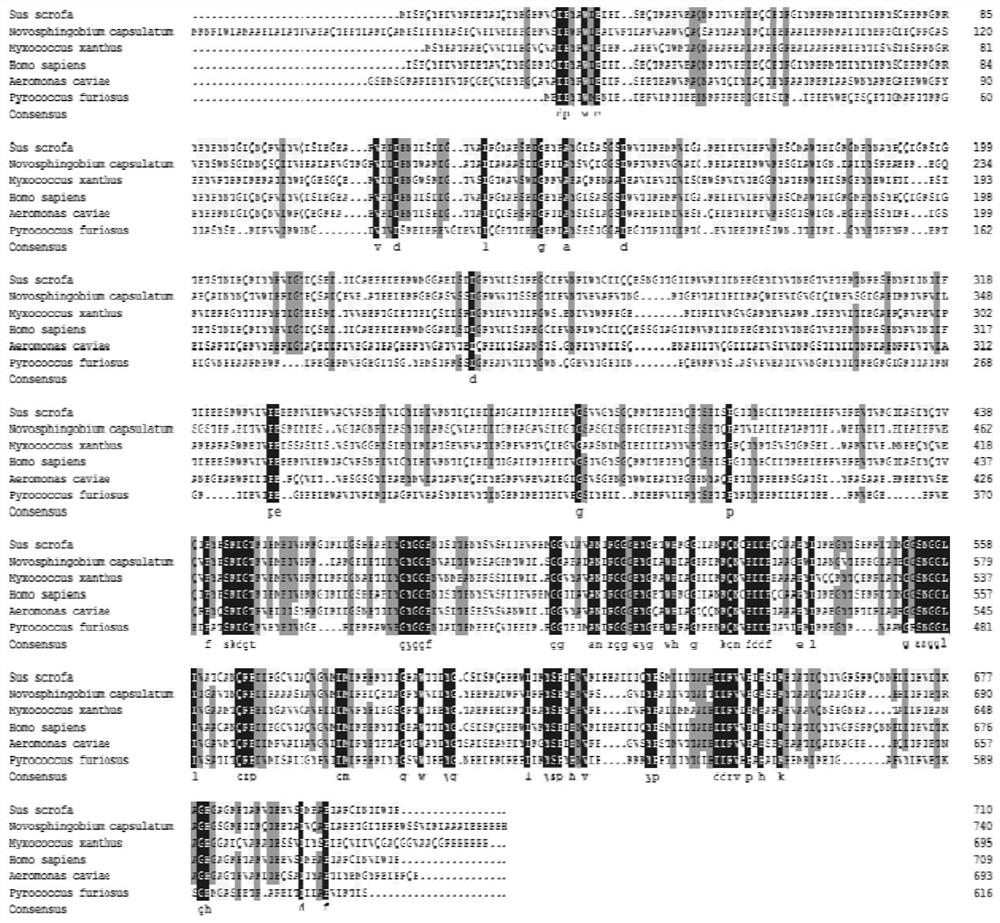
[0034] 2. Search the amino acid sequence of the crystal structure of prolyl endopeptidase in the protein database (PDB database), and use the BLAST (local sequence alignment search basic tool) of the NCBI database to sequence the amino acid sequence of these prolyl endopeptidase Compare, the result is as figure 1 shown.
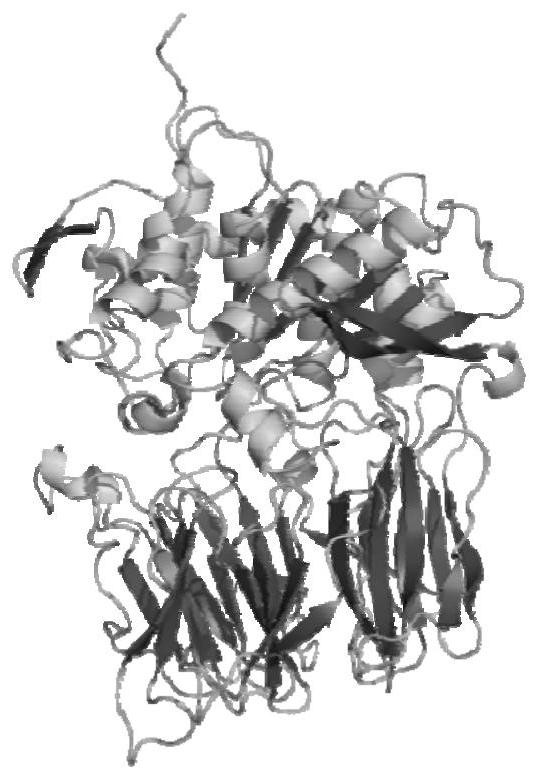
[0035] 3. Select the X-ray crystal structure (PDB ID: 2BKL) of Myxococcusxanthus prolyl endopeptidase (43.84% sequence identity) with the high similarity of Sphaerobacter thermophiles prolyl endopeptidase as template, us...
Embodiment 2
[0038] Example 2: Virtual Saturation Mutation of Residues Surrounding Prolyl Endopeptidase Substrates
[0039] Based on the stable enzyme-substrate complex structure obtained in Example 1, saturation mutations were performed around the substrate. Around P(Pro, proline) in the substrate sequence through the leap module of AMBER18 The 25 residues of prolyl endopeptidase in the range were subjected to virtual saturation mutation, and 575 virtual single-point mutants were generated. Virtual screening was carried out for all the above-mentioned virtual single-point mutants, and 9 with strong binding free energy were selected. a virtual single point mutant. The steps for virtual screening are as follows:
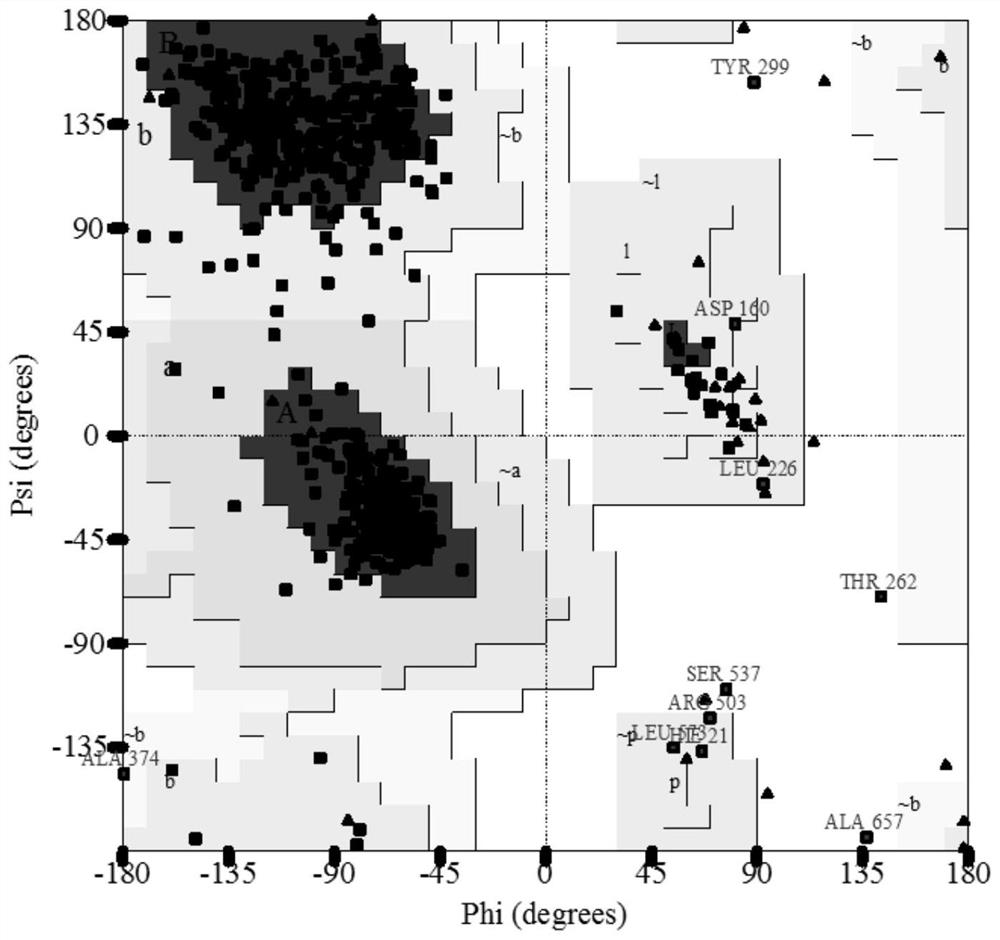
[0040] Constrained dynamics simulations were performed on all mutants to optimize their structures, even though the distance between the C atom of Pro in the substrate sequence and the O atom of Ser537 in the prolyl endopeptidase sequence was kept at The harmonic force consta...
Embodiment 3
[0054] Example 3: Molecular construction and expression of recombinant prolyl endopeptidase and site-directed mutants
[0055] The primer fragment names and sequences used in this embodiment are shown in the following table:
[0056]
[0057] Molecular Construction and Expression of Recombinant Prolyl Endopeptidase
[0058] 1. The Sphaerobacter thermophiles strain was cultured overnight at 37°C in 10 mL LB liquid medium. Use the bacterial genome DNA extraction kit to extract the genome of the Sphaerobacter thermophiles strain, use this as a template, and use P1 and P2 primers to obtain the prolyl endopeptidase target gene fragment (UniProtKB: D1C7Y4) by PCR amplification, the C-terminal of the target gene Containing His-tag, the pET-28a(+) plasmid was digested with EcoRI and NcoI restriction endonucleases respectively, and the recombinant DNA was obtained by reacting with T4 ligase in a metal bath at 16°C for 15 hours. Prolyl endopeptidase target gene PCR identification r...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com