Combinations of miRNA markers, kits and methods for detecting breast cancer
A marker, breast cancer technology, applied in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0099]
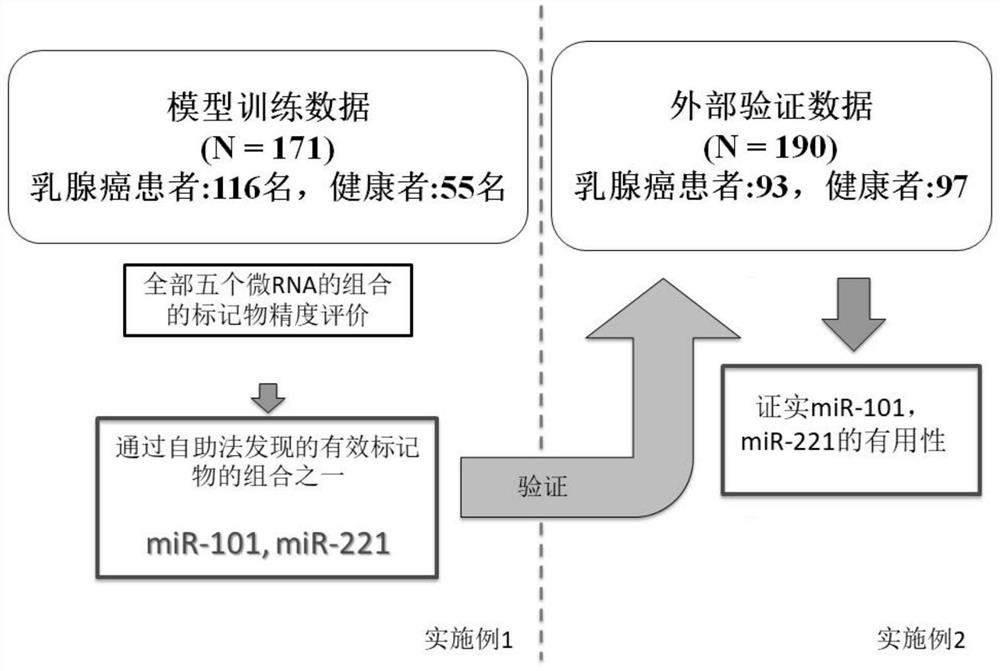
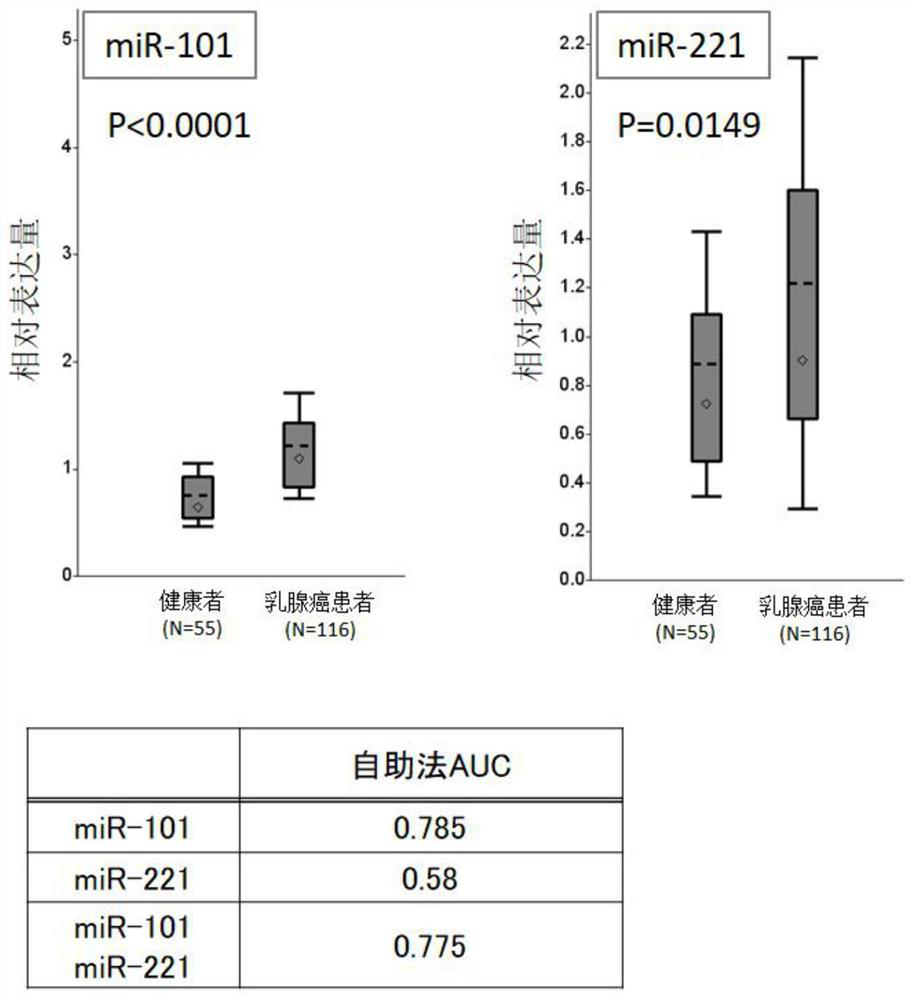
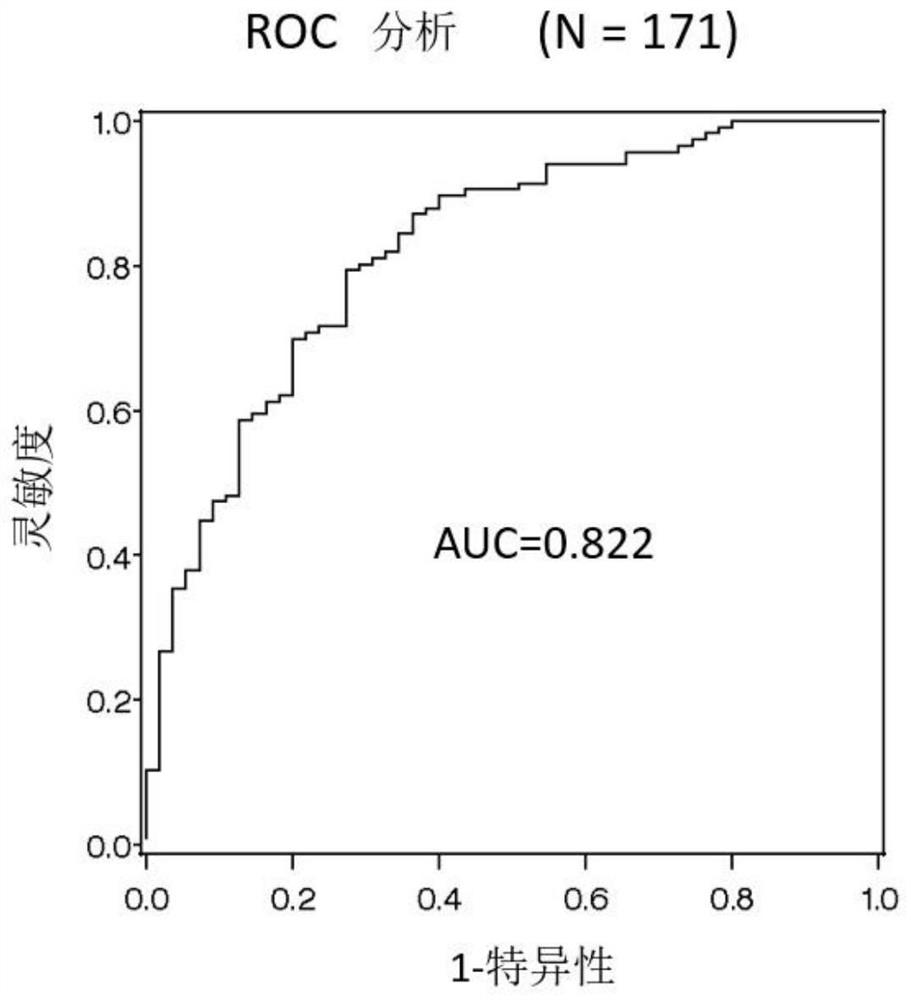
[0100] Five microRNAs (miR-21, miR-29a, miR-101, miR-221, and miR-155) implicated in association with breast cancer were evaluated in detail by the training group[ figure 1 ]. In the training set, the improvement of the marker properties achieved by using the most effective markers of these five microRNAs and their combinations was discussed, and thus the analysis using the bootstrapping method was performed. Quantitative analysis of each microRNA used the quantitative PCR method by reverse transcription reaction. The details of the experiment are described below.
[0101] plasma separation : Use a 5mL or 7mL capacity EDTA-2Na vacuum blood collection tube to collect venous blood from breast cancer patients and healthy subjects, and immediately use it for centrifugation (2500×g / 10 minutes) at room temperature after inversion mixing. Then, the upper plasma fraction was dispensed into another tube with a disposable pipette, and stored at -80°C until use.
[0102] ...
Embodiment 2
[0150]
[0151] With respect to the evaluation markers performed in the training group of Example 1, the discrimination accuracy of each marker was evaluated based on AUC using logistic regression analysis for a validation group consisting of another cohort. Here, miR-101 and miR-221 were selected from the evaluation markers performed in the training group, and the discrimination accuracy of each marker was evaluated based on AUC using logistic regression analysis. Regarding the experimental method, according to Example 2, the microRNA in the purified plasma was used to carry out reverse transcription reaction and quantitative PCR method. Details are described below.
[0152] plasma separation : Use a 5mL or 7mL capacity EDTA-2Na vacuum blood collection tube to collect venous blood from breast cancer patients and healthy subjects, and immediately use it for centrifugation (2500×g / 10 minutes) at room temperature after inversion mixing. Then, the upper plasma fraction was d...
Embodiment 3
[0183]
[0184] With respect to the evaluation markers performed in the training group of Example 1, the discrimination accuracy of each marker was evaluated based on AUC using logistic regression analysis for a validation group consisting of another cohort. Here, miR-101 and miR-155 were selected from the evaluation markers performed in the training group, and the discrimination accuracy of each marker was evaluated based on AUC using logistic regression analysis. Regarding the experimental method, according to Example 3, the microRNA in the purified plasma was used to carry out reverse transcription reaction and quantitative PCR method. Details are described below.
[0185] plasma separation : Use a 5mL or 7mL capacity EDTA-2Na vacuum blood collection tube to collect venous blood from breast cancer patients and healthy subjects, and immediately use it for centrifugation (2500×g / 10 minutes) at room temperature after inversion mixing. Then, the upper plasma fraction was d...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com