Multi-gene family identification and evolution analysis method
A gene family and evolutionary analysis technology, applied in the field of biological information analysis, can solve the problems of limited analysis range, high cost, cumbersome operation, etc., and achieve the effect of mature analysis method, high accuracy and high efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
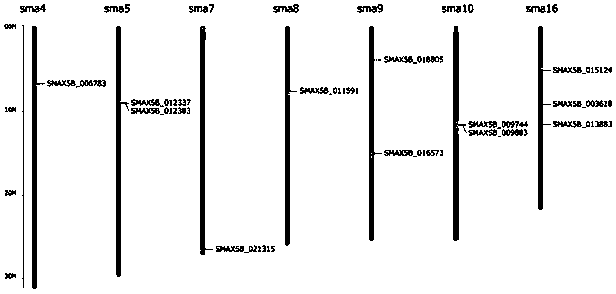
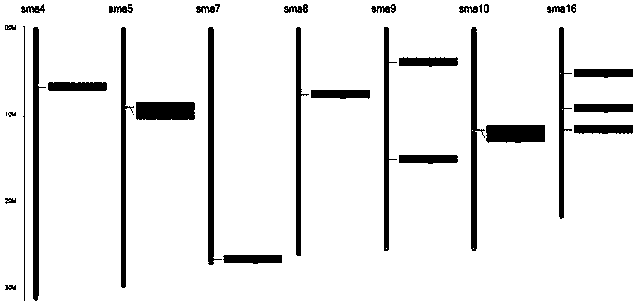
[0063] This example provides a cross-species, multi-dimensional, comprehensive multi-gene family identification and The method of evolutionary analysis, its technological process is as follows figure 1 shown, including the following steps:
[0064] Step 1, statistical gene sequence or protein sequence information;
[0065] Obtain the gene sequence or protein sequence information of the SMAD protein gene family of three species of turbot (Scophthalmus maximus), zebrafish (Danio rerio), and flounder (Paralichthys olivaceus).
[0066] Step 2, carry out identification and evolution analysis for the target species;
[0067] Step 2.1, protein gene family identification
[0068] (1) Compare the gene sequence of the SMAD gene family with the turbot genome, and analyze the structural annotation information of the gene, such as the location of the gene on the chromosome, the CDS sequence of the gene, exons, introns and other information.
[0069] (2) Translating the CDS sequence of ...
Embodiment 2
[0105] In this example, three fish species, turbot (Scophthalmus maximus), zebrafish (Danio rerio), and flounder (Paralichthys olivaceus), are taken as examples to provide a cross-species, multi-dimensional, comprehensive target species and close relatives. A method for identification and evolution analysis of multi-gene families of species, the technological process of which is as follows figure 1 shown, including the following steps:
[0106] Step 1, identifying protein gene families for related species;
[0107] The following is an analysis of the SMAD protein gene family of turbot (Scophthalmus maximus, sma) and its close relatives, zebrafish (Daniorerio, dre) and flounder (Paralichthys olivaceus, pol).
[0108] According to the reference genome information of the two species of zebrafish and flounder and their respective SMAD protein gene family information, the SMAD protein information and gene sequences of zebrafish and flounder species were analyzed respectively by us...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com