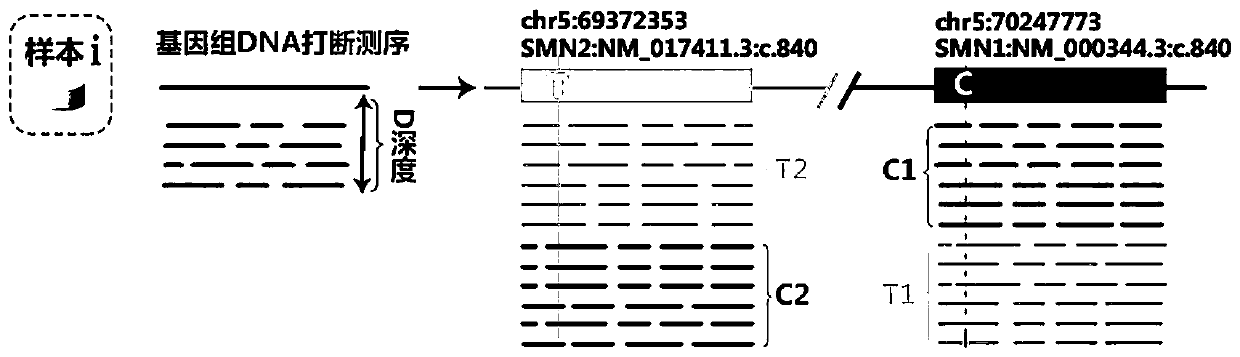
Method and system for detecting SMN1 gene mutation by means of high-throughput sequencing
A sequencing and gene technology, applied in the fields of genomics, biochemical equipment and methods, computer-aided medical procedures, etc., can solve the problem of inappropriate SMA molecular diagnosis, and achieve the effect of high detection rate and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0074] Example 1. Detection of SMN1 Homozygous Deletion in Subjects Clinically Diagnosed as SMA
[0075] This example involves using the detection system of the present invention to detect homozygous deletion of SMN1 in subjects clinically diagnosed as SMA.
[0076] Subject selection
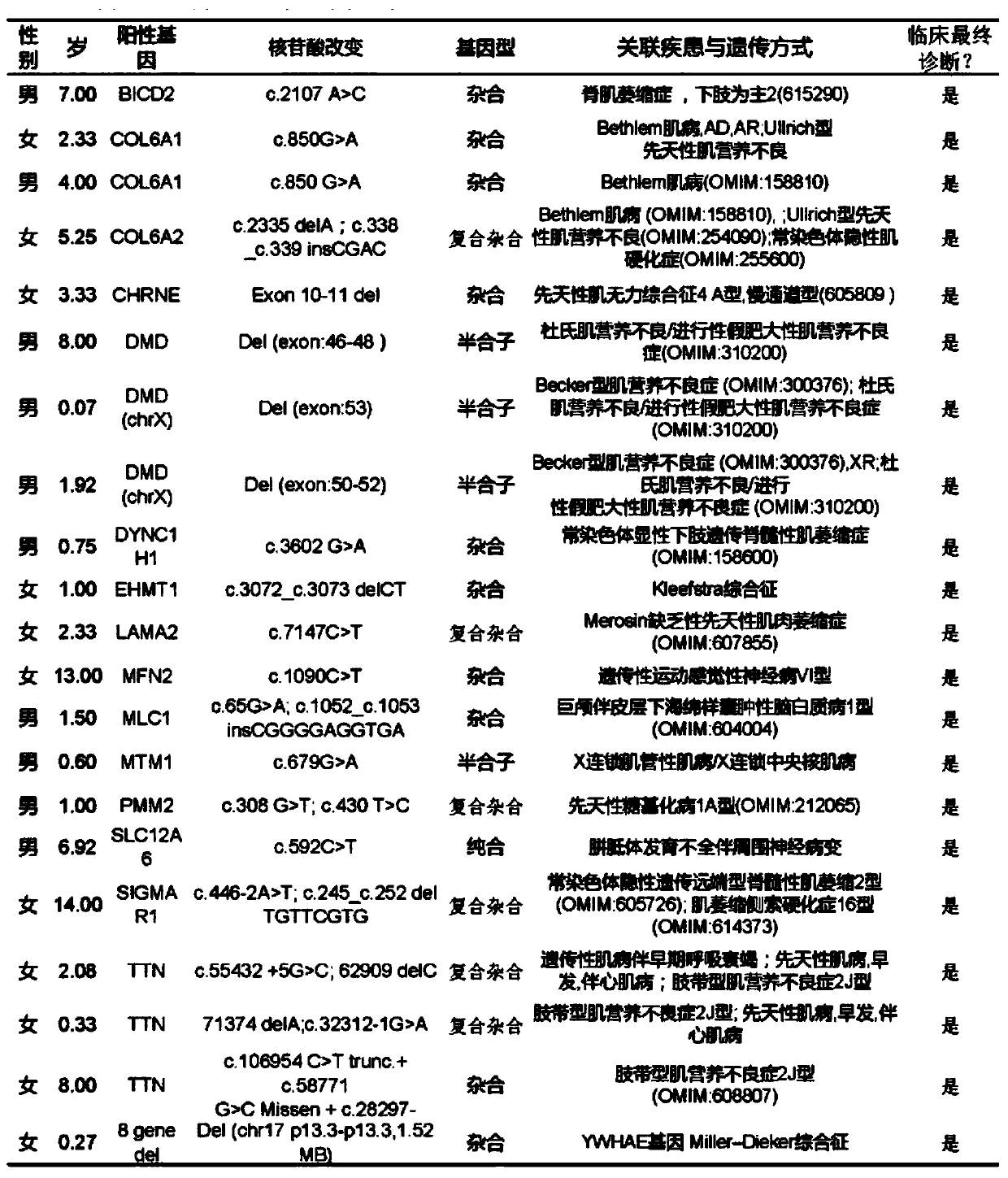
[0077] The subjects in this example were patients who visited the hospital from June 2015 to July 2018, and peripheral whole blood biological samples were obtained from these patients during the visit. The enrolled cases all had the characteristic phenotype of neuromuscular disease, and the physicians who submitted for examination provided a description of the clinical characteristics and various special examination results corresponding to each patient (the private information such as the patient's name has been hidden). For the use of sample test results for clinical research and data publication, written informed consent has been obtained from patients or guardians and family members par...
Embodiment 2
[0090] Example 2. Differential diagnosis for subjects with non-SMN1 homozygous deletion
[0091] Adopt the general method of whole exome to detect genetic disease to analyze whether subjects (especially 118 subjects identified as non-SMN1 homozygous deletion in embodiment 1) have other neuromuscular diseases similar to SMA phenotype, for a differential diagnosis.
[0092] The concrete steps of described general method comprise:
[0093] 1) Raw data yield statistics: Remove adapter contamination, filter out reads with an average quality value lower than 20, and filter out bases with a quality value lower than 20 from the end of the reads.
[0094] 2) Alignment: The comparison statistics between the data and the reference sequence (comparison software BWA), the reference genome adopts the hg19 genome.
[0095] 3) Mutation detection: Use GATK to compare the results for alignment rearrangement and quality correction, and then use GATK's HaplotypeCaller algorithm to call mutati...
Embodiment 3
[0104] Example 3. Detection of SMN1 Homozygous Deletion in Subjects Clinically Suspected of SMA
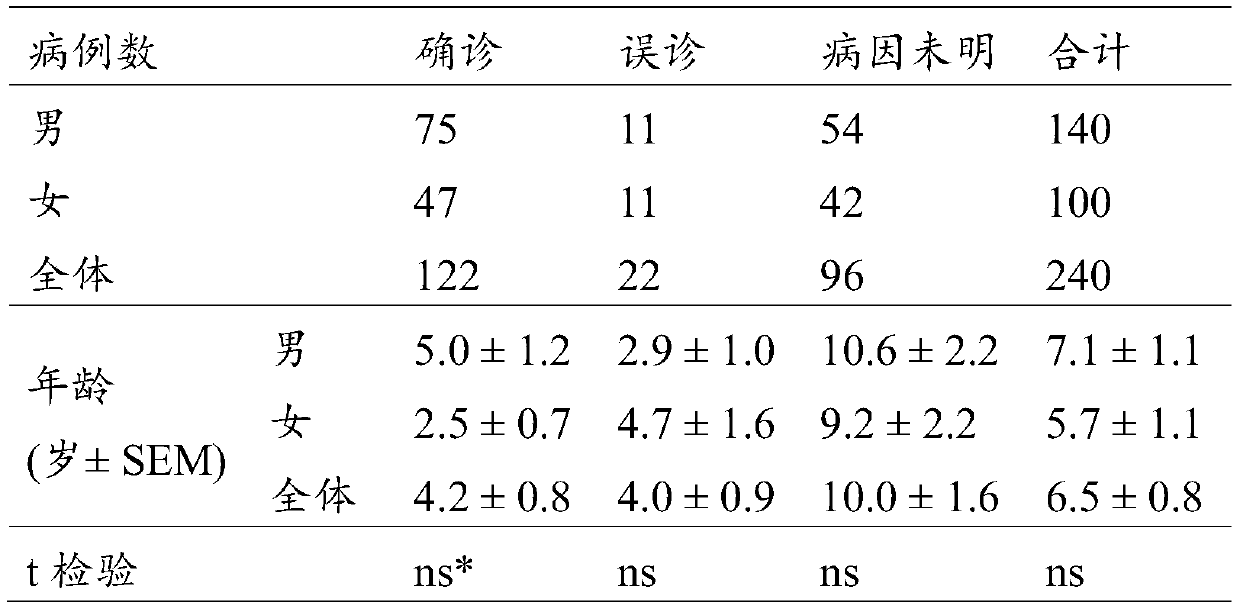
[0105] As of August 2018, among all the subjects who had the clinical features of neuromuscular disease but were not suspected of SMA in the initial diagnosis, for the subjects whose genetic pathogenic factors could not be ruled out, as in Example 1, the inventors The detection means of the present invention described above detected 56 subjects carrying SMN1 homozygous deletion mutations (see Table 2).
[0106] Table 2
[0107]
[0108] *ns: not significant
[0109] Based on the results obtained in Examples 1 to 3, it can be seen that the method of the present invention can provide the following comprehensive diagnostic information for subjects with different conditions.
[0110] 1) Differential diagnosis was carried out on 240 patients who were preliminarily judged as SMA:
[0111] A. 122 confirmed cases of SMA (122 / 240, 50.8%): Among all 240 patients who were preliminarily...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com