Fusion protein of monomeric streptavidin and Gaussian luciferase and its application
A technology of streptavidin and fusion protein, applied in the field of fusion protein of monomer streptavidin and Gaussian luciferase, can solve the problems of substrate aggregation, time-consuming, cumbersome process, etc. The effect of simple production and production process and broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0099] Embodiment 1: Construction of fusion expression vector
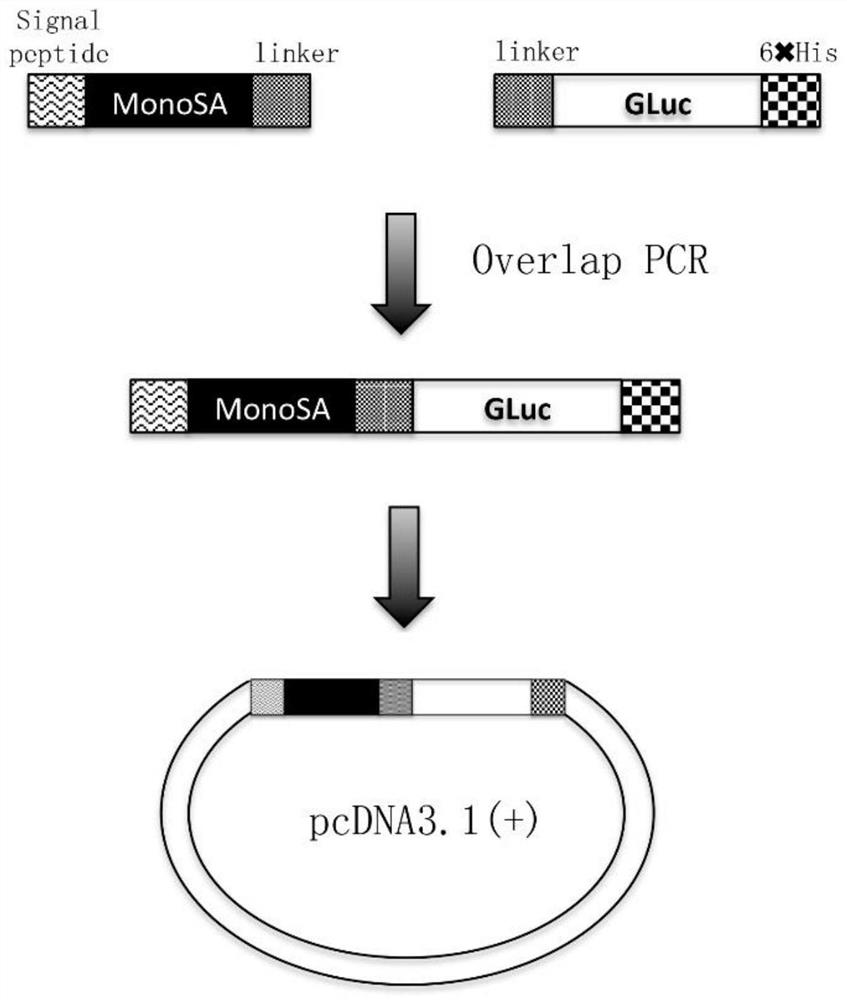
[0100] Such as figure 1 As shown, the MonoSA gene and the Gaussian luciferase gene were used as templates to design and synthesize primers for PCR amplification to obtain the MonoSA and Gaussian luciferase gene amplification products respectively. Based on this PCR product, a second round of bridging PCR was performed to obtain the amplified product of the fusion gene of MonoSA and Gaussian luciferase (sequence such as SEQ ID NO: 13). The PCR amplified product was detected by electrophoresis on 1% agarose gel, and the product was recovered with a gel recovery kit. The recovered product and the vector plasmid pcDNA3.1(+) were subjected to a double enzyme digestion reaction with EcoR I and Hind III respectively. After the reaction was completed, electrophoresis was performed on a 1% agarose gel for detection, and the product was recovered with a gel recovery kit . The products recovered from PCR digestion and the...
Embodiment 2
[0102] Example 2: Transfection, expression and purification of fusion expression vector
[0103] Mix 25 μg of the constructed pcDNA3.1-MonoSA-Gluc fusion expression plasmid with 1 ml of cell culture medium evenly. Another 920 μL cell culture medium was mixed with 80 μL lipo2000 transfection reagent (invitrogen). Mix the diluted expression vector and the diluted transfection reagent, and let stand at room temperature for 20 minutes. Prepare 25ml concentration of 2×10 6 / ml cells, add the mixed pcDNA3.1-MonoSA-Gluc fusion expression plasmid and transfection reagent mixture into CHO cells, and shake gently. 37°C, 8% CO 2 conditions for 3 days.
[0104] Collect the cell culture fluid, centrifuge at 6000rpm for 10min, and collect the supernatant. After the supernatant was filtered, it was passed on to a nickel column, and 10 ml of PBS was equilibrated. Then wash 10 ml with buffer (20mM Tris, pH 8.0, 150mM NaCl, 20mM imidazole) to remove foreign proteins, and finally elute wit...
Embodiment 3
[0106] Example 3: Fusion protein luciferase bioluminescent detection
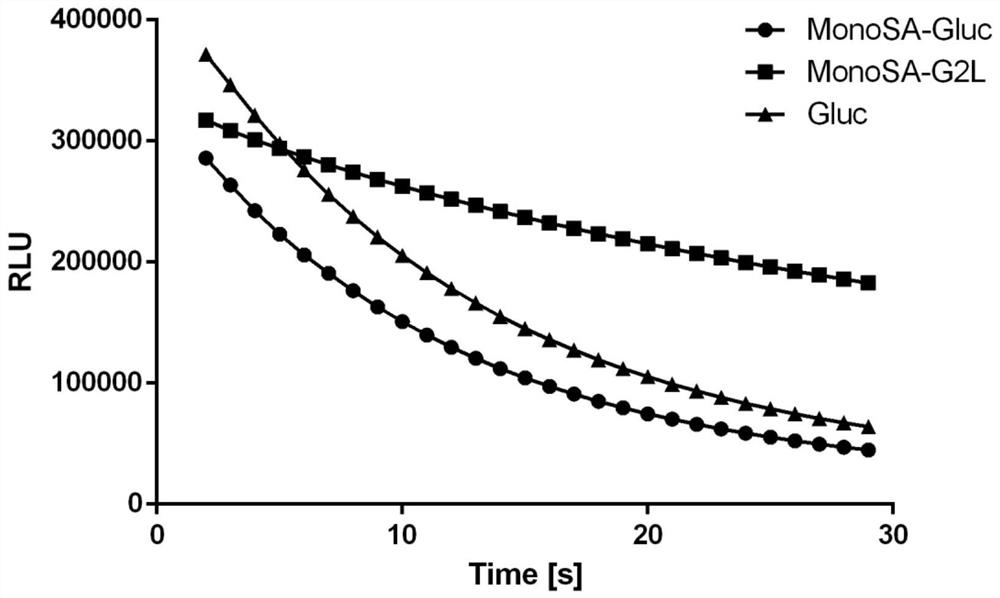
[0107] MonoSA-Gluc fusion protein and MonoSA-G2L fusion protein were diluted to 3nM with substrate diluent (50mM Tris, pH 8.0, 100mM NaCl), respectively, and 10 μL was added to a 96-well microtiter plate. Then add 90 μL of the substrate coelenterazine diluted to 10 μM with the same solution, and read the luminous intensity with the self-luminescence module of the microplate reader, and the results are as follows image 3 As shown, the luminescence intensity of wild-type MonoSA-Gluc fusion protein is equivalent to that of Gluc protein at the same concentration. The luminescence intensity of the mutant fusion protein MonoSA-G2L fusion protein was comparable to that of Gluc, but it obviously showed continuous luminescence characteristics, and the luminescence did not weaken significantly after 30s.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com