Primer probe set for detecting novel corona virus SARS-CoV-2 and detection method
A primer-probe, coronavirus technology, used in biochemical equipment and methods, DNA/RNA fragments, recombinant DNA technology, etc., can solve the problem of sample backlog of suspected cases, inability to obtain results quickly, and detection application limitations, etc. Short detection time, overcoming long detection time and good repeatability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0054] The primary screening of embodiment 1 specific primer probe combination
[0055] The full sequence of the new coronavirus SARS-CoV-2 announced by the Chinese Center for Disease Control and Prevention contains 10 genes for comprehensive analysis and comparison, combined with the needs of epidemic prevention and control, to achieve the purpose of rapid screening, the core solution of the present invention is fast, sensitive, Accurate these three basic points, select the most conservative ORF 1ab gene sequence from 10 genes to design primer probes, and in order to detect the new coronavirus SARS-CoV-2 faster and more accurately, targeting the ORF 1ab gene Perform single gene detection, reduce the interference of detection of multiple genes, improve the amplification speed and detection sensitivity;
[0056] The specific design uses DNAMAN software for coronavirus homology analysis and b1ast sequence analysis, and screens out two intervals with excellent conservation and sp...
Embodiment 2
[0075] The determination of embodiment 2 specific primer probe combinations
[0076] On the basis of the 5 primer probe combinations in the above-mentioned embodiment 1, further adaptive screening is carried out with the nucleic acid of the new coronavirus SARS-CoV-2 strain culture, and the selection of a pair of primer probes with the highest sensitivity is provided by the present invention. The primer probe set;
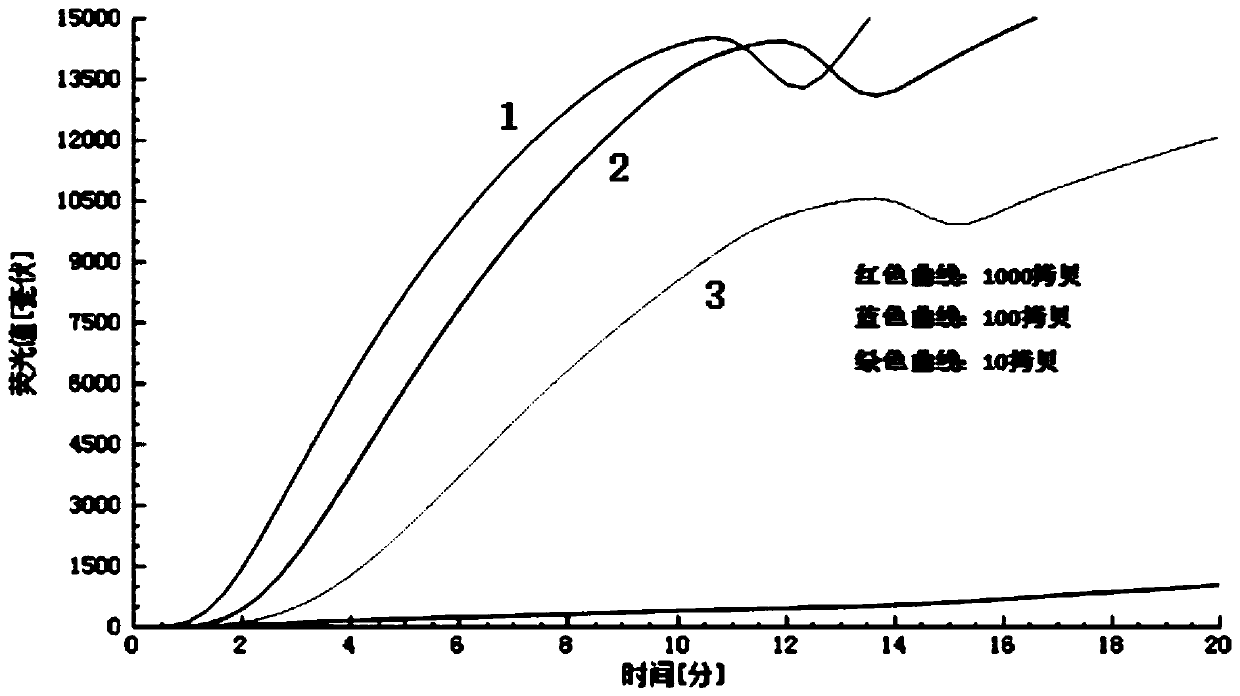
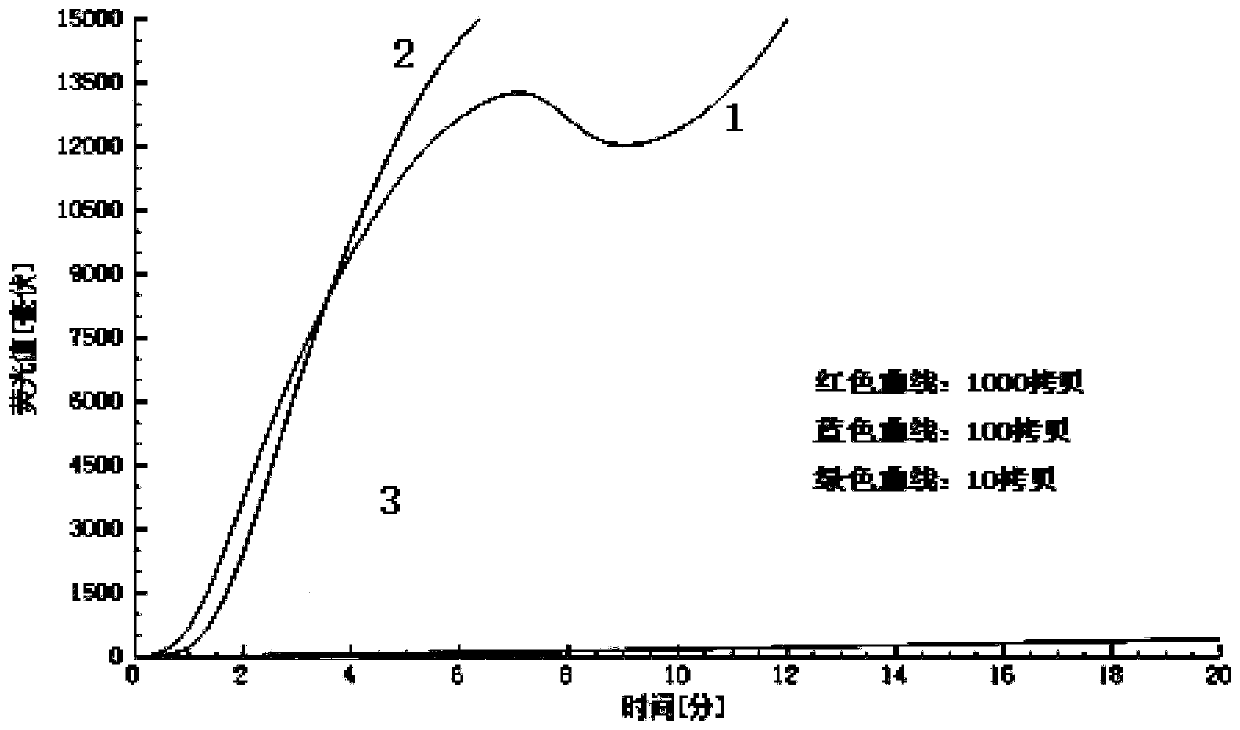
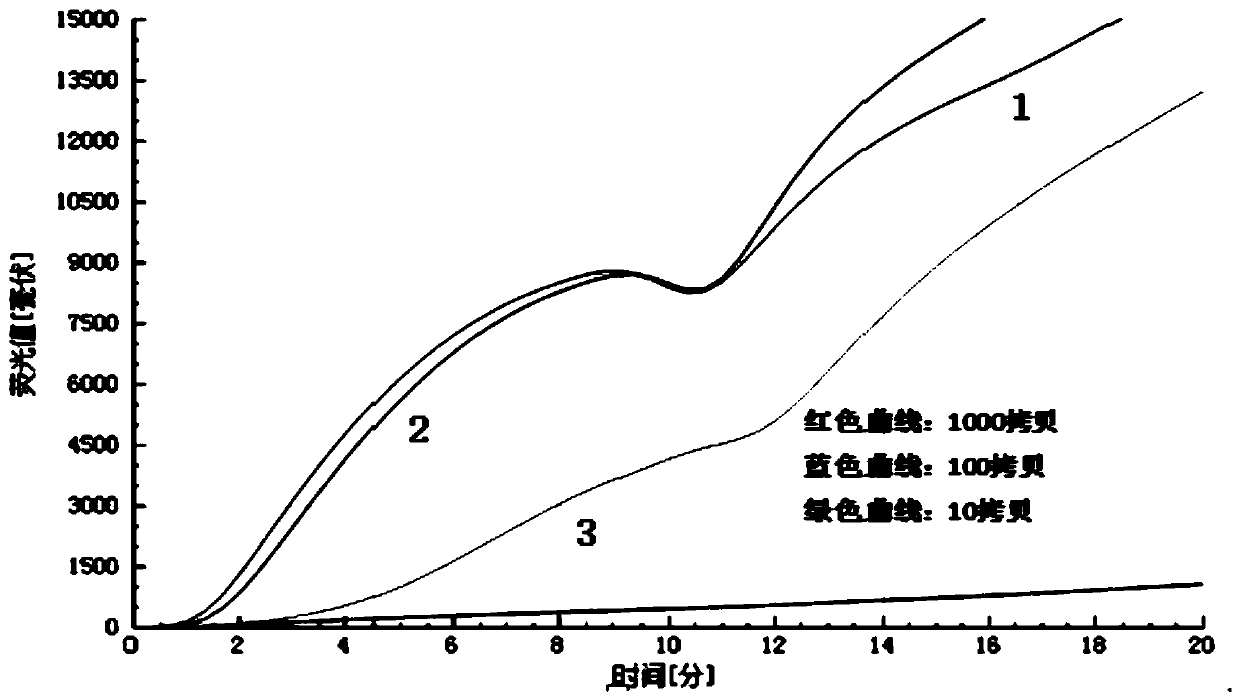
[0077] The purpose of this embodiment is to screen out the most sensitive primer probe group, and the nucleic acid stock solution is diluted to 10 times by 10 times. 2 、10 3 、10 4 、10 5 、10 6 、10 7 、10 8 、10 9 、10 10 There are ten gradients in total, and the sample volume is 5 μL.
[0078] The specific implementation method is as follows: (1) prepare 5 pairs of primer probe set solutions according to the concentration of probe 0.02mM and primer 0.03mM, prepare the diluted nucleic acid sample and the RT- The RAA fluorescence basic kit is ready for use, and...
Embodiment 3
[0112] Example 3 Sensitivity Verification of Detecting Novel Coronavirus Kit
[0113] The novel coronavirus detection kit provided by the present invention contains two parts: a reaction unit and a reaction buffer; the reaction buffer contains the novel coronavirus SARS-CoV-2 specific primer probe set determined in Example 2.
[0114] The reaction buffer is prepared by adding the specific primer probe set described in this application to the general reaction buffer in the F00R01 kit provided by Jiangsu Qitian Gene Biotechnology Co., Ltd. The components in the reaction buffer are: : upstream primer, downstream primer, probe, Tris-HCl buffer, MgAc and PEG 20000;
[0115] The reaction unit is the general reaction unit in the kit provided by Jiangsu Qitian Gene Biotechnology Co., Ltd. with the product number F00R01. It mainly includes the following components: ATP, dNTPs, recombinase, single-strand binding protein, exonuclease, DNA polymerase , reverse transcriptase.
[0116] Us...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com