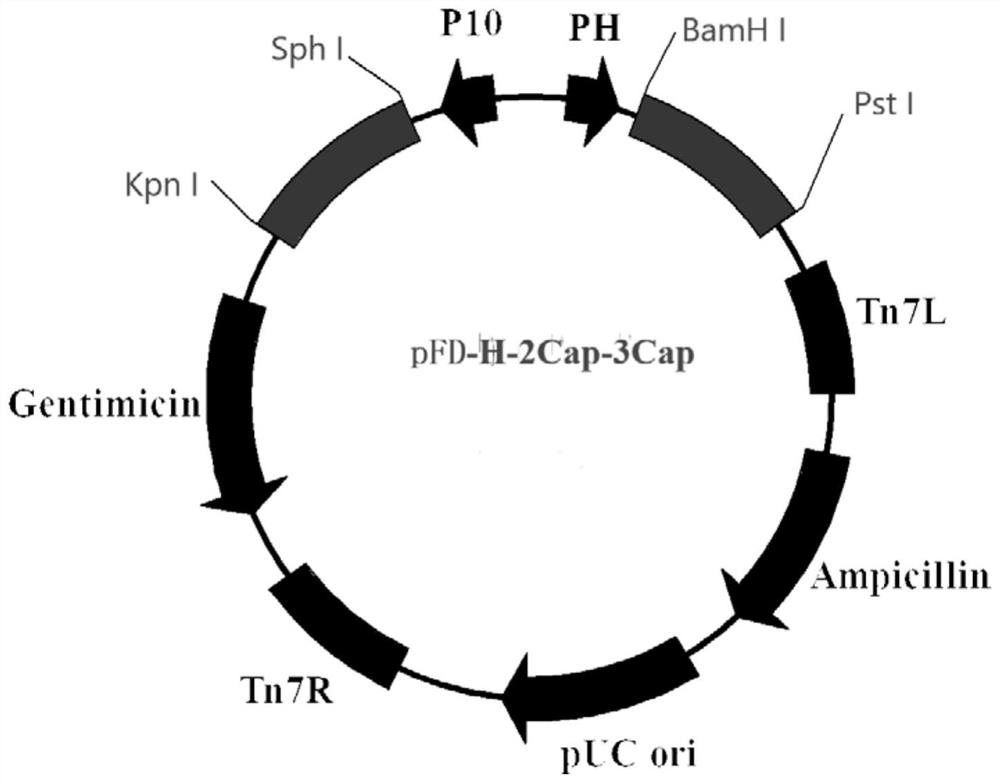
A method for efficiently expressing pcv2cap and pcv3cap fusion proteins
A fusion protein and protein technology, applied in the field of molecular biology, can solve the problems of complicated purification process and low expression amount of fusion protein, and achieve the effects of high biological activity, high expression efficiency and lower production cost.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Example 1 Construction of a rodovirus
[0041] 1. The result of the gene
[0042] Referring to "Establishment of Porcine Ring Virus 2 Sybr Green I Real Time Fluorescent Quantitative PCR Detection Method", "Establishment of Porcine Ring Virus 3 Sybr Green I Real Time Fluorescent Quantitative PCR Detection Method" 2 and 3 The plasmid of the porcine circular virus genome PEASY-BLUNT-PCV2, PEASIY-BLUNT-PCV3, and preserved for -80 ° C.
[0043] Table 1 Purpose Gene PCR primer sequence
[0044] .
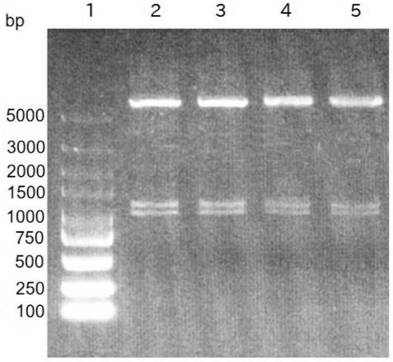
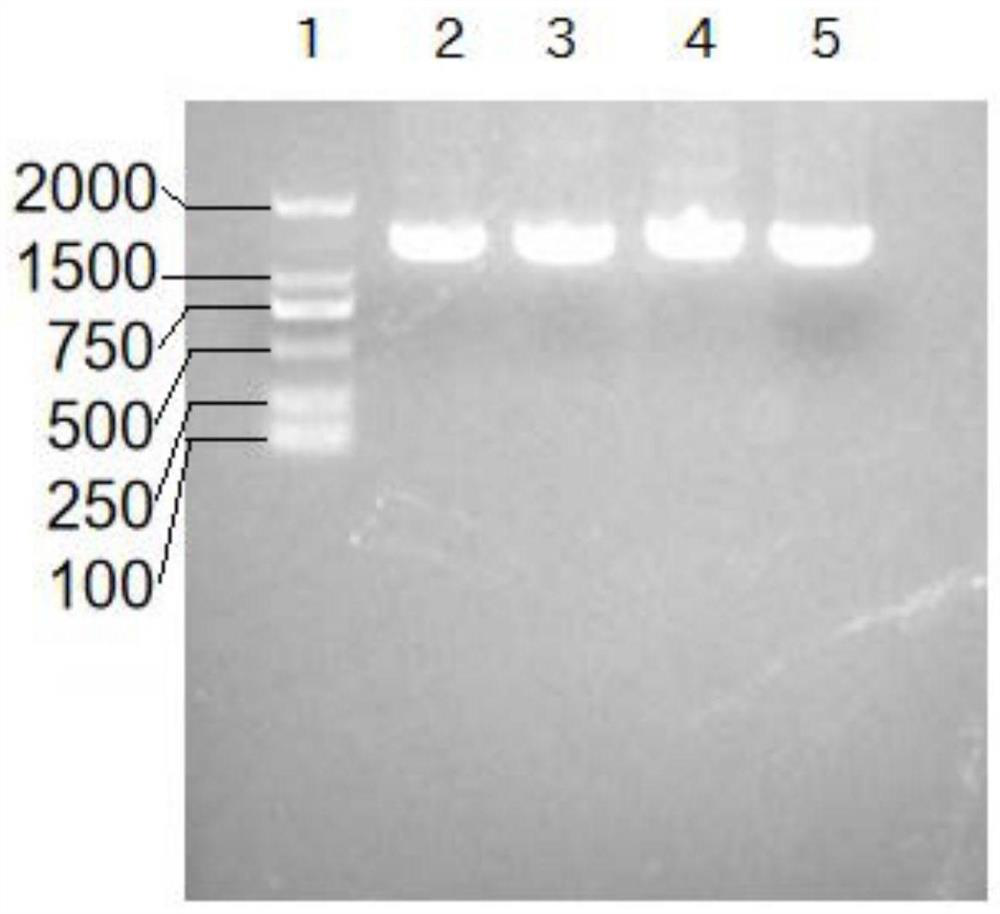
[0045] According to Table 1 synthesis primers, PEASY-BLUNT-PCV2 is a template, and the amplification of PCV2 CAP base sequences is performed in HBM-PCV2CAP F, Linker1-PCV2CAP R, and the amplified product is recovered by glue recovery kit. Named 2CAP. The PEASY-BLUNT-PCV3 is a template, and the amplification of PCV3 CAP base sequences in LINKER2-PCV3CAP-PHF, PCV3cap-pHR is primer, and the amplified product is recovered with a glue recovery kit and is named 3 cp. The recombinant sequen...
Embodiment 2
[0080] Example 2 Expression of fusion protein
[0081] P2 recombinant baculoviruses were inoculated into MOI = 1, 0.5, 0.1 viral volume to 2.5 × 10 6 A / MLSF9 cells. 200 μl of the supernatant was charged every 24 hours, and 5 days were collected, and the protein sample was purified, the steps are as follows:
[0082] 1) The collected supernatant 4 ° C, 5000 × g is centrifuged for 30 min to remove cell debris and impurities, and the cultured supernatant is collected;
[0083] 2) Centrifugal cells were centrifuged at 30000 rpm 1 h, harvested precipitation, ie sterile PBS resuscitation;
[0084] 3) Sterile PBS resuspended precipitation to the upper level liquid surface of 10% -30% -50% sucrose density gradient, 35 000 rpm centrifugation 1.5 h for purification, carefully collect white floc between 30% -50% sucrose layer The strip was resuspended with PBS, 30 000 rpm, centrifuged 1.5 h removal of sucrose, collected, 2 ml sterile PBS resuspended, -80 ° C preservation. SDS-PAGE electrop...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com