Fc-gamma receptor mutants
A mutant and receptor technology, applied in the direction of antibody mimics/stents, allergic diseases, drug combinations, etc., can solve the problems of prolonging serum half-life, short half-life, short residence time, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
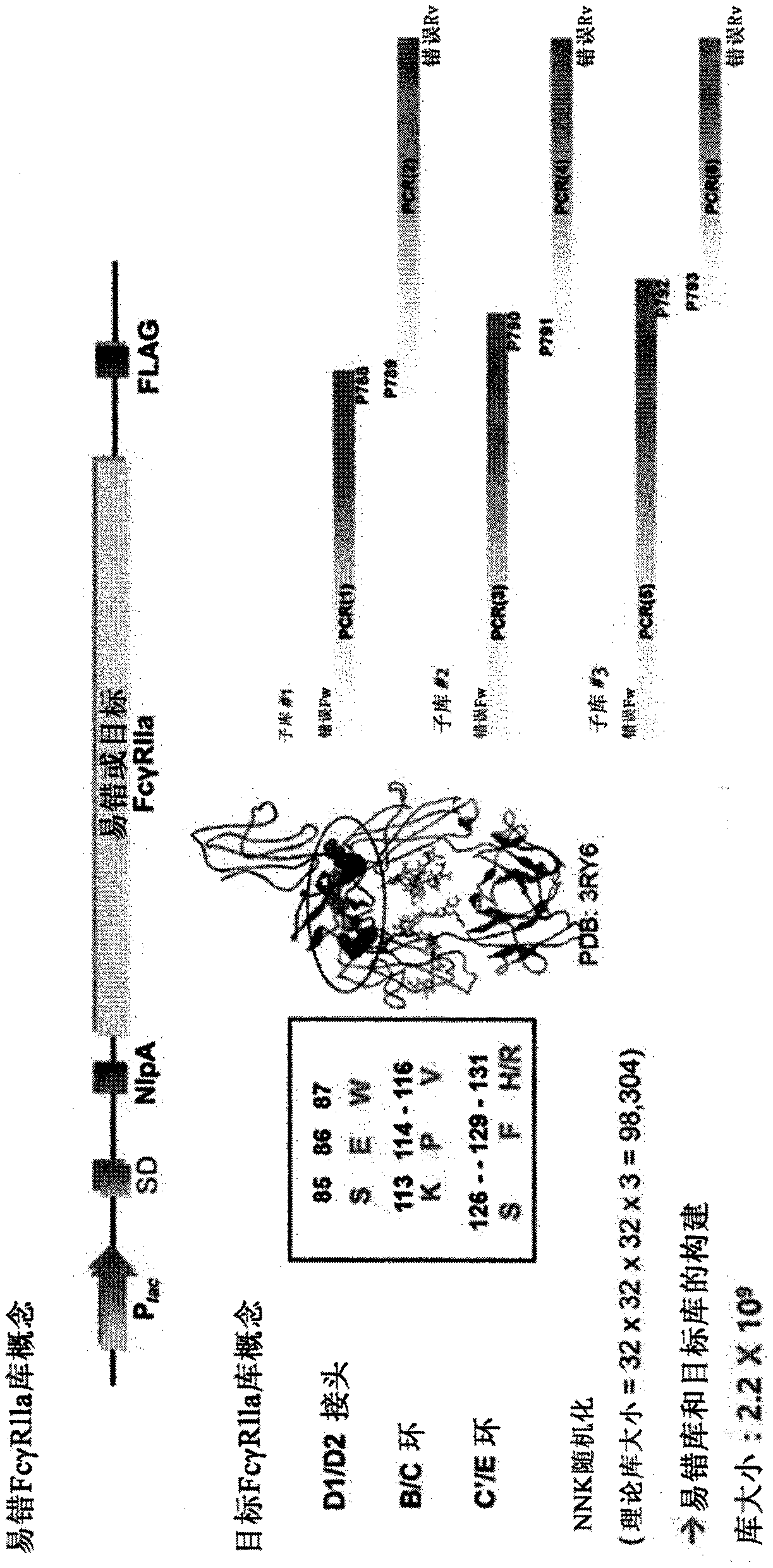
[0147] Example 1. Construction of FcγRIIa mutant library
[0148] Based on the pMopac12-NlpA-FcγRIIa-FLAG vector, the FcγRIIa gene introducing random mutations was prepared using MJ#160 and MJ#161 primers so that about 0.2% of random mutations occurred in FcγRIIa. In addition, target inserts incorporating various amino acids were prepared by introducing a degenerate codon NNK at the IgG Fc binding site of FcγRIIa using MJ#160, MJ#161, p788, p789, p790, p791, p792, and p793 primers (Table 1). The prepared two inserts were treated with SfiI (New England Biolab) restriction endonuclease and ligated to the vector. Then, by transforming into Escherichia coli (E.coli) Jude1 ((F'[Tn10(Tet r )proAB + lacI q Δ(lacZ)M15]mcrAΔ(mrr-hsdRMS-mcrBC)Φ80dlacZΔM15ΔlacX74 deoR recA1 araD139Δ(ara leu)7697 galUgalKrpsLendA1nupG) to construct FcγRIIa mutant library (library size: 2.2×10 9 )( figure 1 ).
[0149] [Table 1]
[0150]
[0151]
[0152]
Embodiment 2
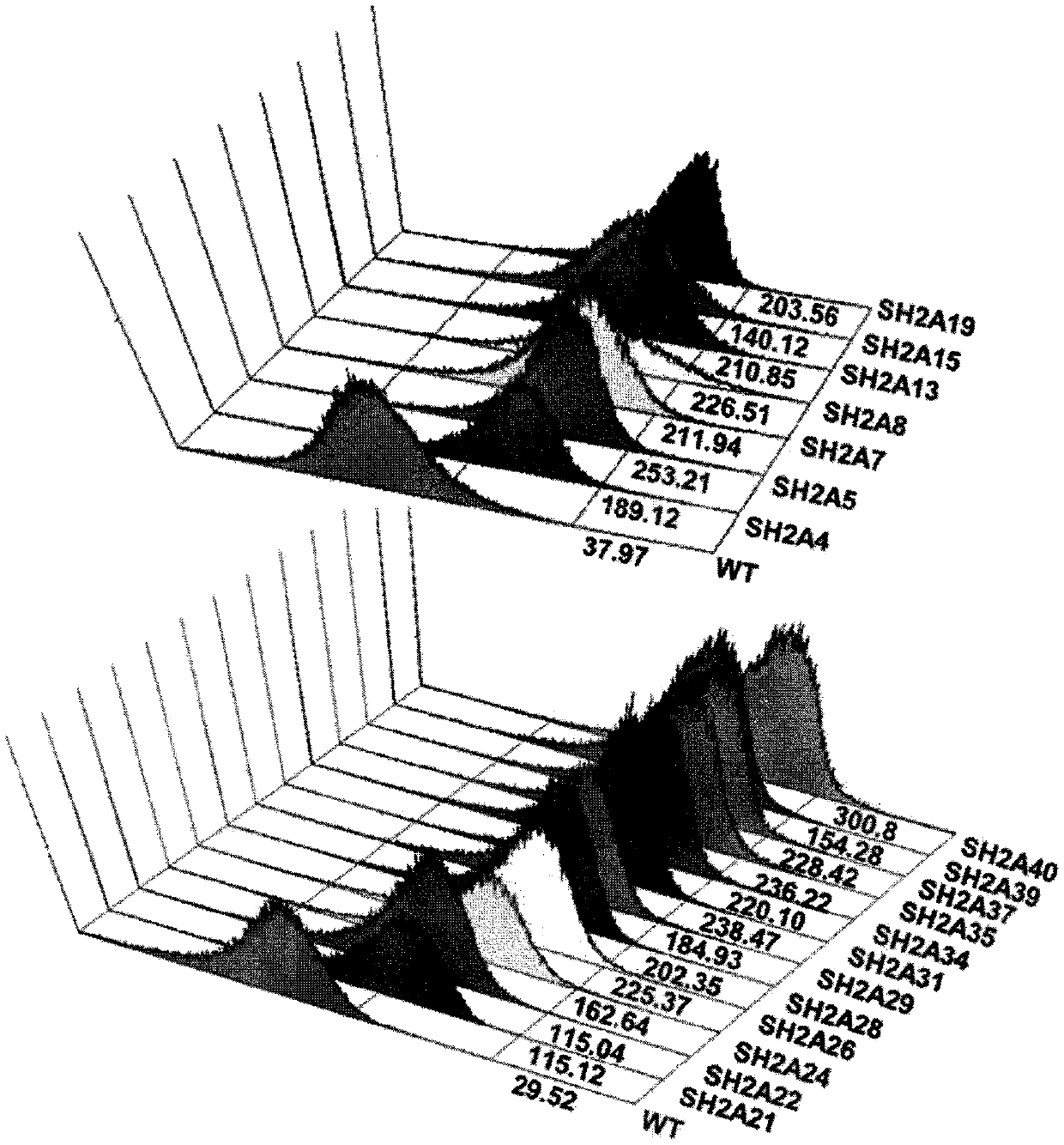
[0153] Example 2. Screening of FcγRIIa mutant library by bacterial culture and flow cytometry
[0154] Culture 1 mL of the established FcγRIIa mutant library cells in Super Broth (TB) medium containing 2 wt / vol% glucose and chloramphenicol (40 μg / mL) at 37°C for 4 hours with shaking at 250 rpm . The cultured pool cells were inoculated into TB medium at 1:100, and then cultured to OD at 37°C while shaking at 250 rpm 600 0.6. Then, after incubation at 25° C. for 20 minutes for cooling, 1 mM isopropyl-1-thio-β-D-galactopyranoside (IPTG) was added to induce expression. After the culture was completed, the cells were recovered, and the OD 600 Normalization Centrifuge at 14000 rpm for 1 min. After collection, cells were resuspended by adding 1 mL of 10 mM Tris-HCl (pH 8.0), and centrifuged for 1 min. This washing process was repeated twice. After resuspension in 1 mL of STE [0.5M sucrose, 10 mM Tris-HCl, 10 mM EDTA (pH 8.0)], the outer cell membrane was removed by spinning at ...
Embodiment 3
[0155] Example 3: Cloning, expression and purification of isolated SH2A40 mutants for expression in mammalian cells
[0156] To express the screened SH2A40 mutants in HEK293F cells, pMAZ-FcγRIIa (wild type) and pMAZ-FcγRIIa mutant (SH2A40) were prepared by PCR amplification of the gene using Vent polymerase and MJ#162 and MJ#163 primers, and then Ligation was performed by restriction enzyme treatment with BssHII and XbaI (New England Biolab). The gene cloned into the mammalian cell expression vector pMAZ was transfected into HEK293F cells and expressed transiently at a scale of 300 mL. After the incubation was complete, the cells were removed by centrifugation at 2000 rpm for 10 minutes, and the supernatant was taken and equilibrated with 25xPBS. The resulting solution was filtered through a 0.2-μm bottle top filter (Merck Millipore). After adding 1 mL of a Ni-NTA agarose (Qiagen) slurry equilibrated with PBS, the solution was stirred at 4°C for 16 hours before being poured ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com