Method for detecting respiratory viruses through next generation sequencing with high flux and application
A second-generation sequencing and detection method technology, which is applied in the field of medical testing, can solve the problems of "false negative" cases and achieve high sensitivity, pollution avoidance and high precision
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0102] Example 1: Take 2019-nCoV as an example
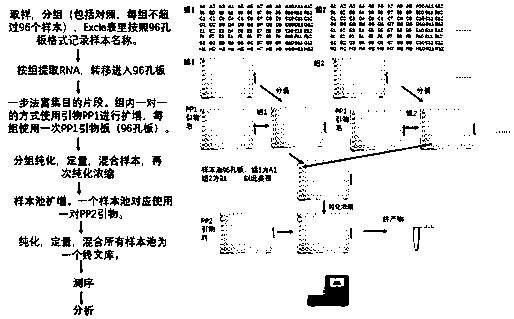
[0103] (1) Collect specimens and group them into groups, with 96 samples in each group (including positive and negative controls), and record the sample names and other information in an Excel table.
[0104]Nasal swab: Insert one swab into the nose and palate, turn it and take it out carefully, take another swab and take the other nostril in the same way. Dip the swab tip into the sampling tube. Then carry out the pretreatment of nucleic acid extraction: cut the swab, put it into a 2.0ml test tube, add it to 400μl PBS, and take 200μl supernatant sample for extraction after vigorous shaking.
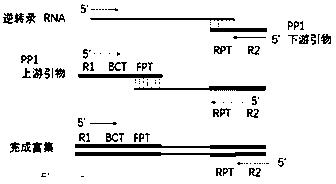
[0105] (2) Extraction of RNA: extract according to the product instructions. Aliquot RNA samples into 96-well plates, 20 μl per well. Then one-step enrichment of nucleic acid target fragments (using SuperScript TM III One-Step RT-PCR System kit as an example):
[0106] (2.1) Set the following program on the PCR instrument.
[0107] ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com