Oligonucleotide and method for detecting ABCG2 gene relative expression quantity in sample
A relative expression level, in-sample technology, applied in the field of detecting the relative expression level of the ABCG2 gene in the sample, can solve the problems of high cost and poor specificity, and achieve the effect of simple operation, high sensitivity, and improved experimental efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0049] The nucleic acid detection method used to detect the relative expression level of ABCG2 in samples can be used to assist in the formulation of lung cancer chemotherapy regimens and the selection of chemotherapy drug doses in clinic, and can also predict the prognosis of patients. The method includes:
[0050] (1) Red blood cell lysate, including 16 μmol / L ammonium chloride, 1 mmol / L potassium bicarbonate and 12.5 μmol / L EDTA.
[0051] (2) RNA extraction reagents, including TRIzol, chloroform, isopropanol, 75% ethanol and RNase-free water.
[0052] (3) RNA reverse transcription reagent, ReverTra Ace qPCR RT Kit kit (TOYOBO company).
[0053] (4) Detection system PCR reaction solution, THNDERBIRD Probe qPCR Mix (2×) (TOYOBO Company). Detect the upstream primer ABCG2-F, downstream primer ABCG2-R and probe ABCG2-Probe of the ABCG2 gene; detect the upstream primer actin-F, downstream primer actin-R and probe actin-Probe of the internal reference gene actin. The primer and...
Embodiment 2
[0062] 1. The operation process of total RNA extraction from peripheral blood samples:
[0063] (1) Collect 1ml of anticoagulated fresh blood, and register age and gender;
[0064] (2) Add 1ml of erythrocyte lysate into a clean 1.5ml centrifuge tube, take 0.5ml of anticoagulated blood and mix well.
[0065] Stand at room temperature for 10 minutes;
[0066] (3) Centrifuge at 5000rpm for 5min, discard the supernatant, and collect the cells at the bottom;
[0067] (4) Add 0.5ml red blood cell lysate again, centrifuge at 5000rpm for 5min, discard the supernatant, and collect the cells at the bottom;
[0068] (5) Add 1ml TRIzol to the cells, pipette repeatedly until the precipitate is completely dissolved, and let stand at room temperature for 5 minutes;
[0069] (6) Add 0.2ml chloroform and shake evenly;
[0070] (7) Centrifuge at 14000rpm at 4°C for 10min, absorb the supernatant layer and transfer it to another new centrifuge tube (do not absorb the white middle layer);
[...
Embodiment 3
[0087] The method of the invention is used to detect the peripheral blood samples of the healthy population.
[0088] Take 24 cases of peripheral blood samples from healthy people for examination, extract RNA according to the method described in Example 2, perform reverse transcription, prepare reagents and perform fluorescence quantitative PCR detection.
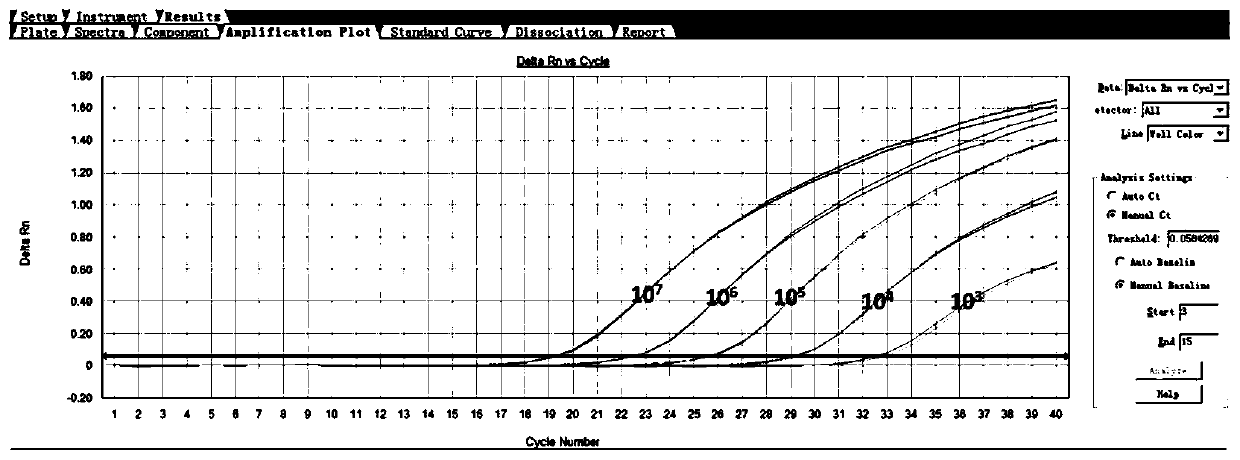
[0089] For each sample, 2 μL of cDNA obtained by reverse transcription was added to the detection system PCR reaction solution. At the same time, make one copy of positive control, negative control and blank control, and two copies of standard curve of internal reference gene / target gene. A 96-well fluorescent PCR instrument can detect 16 samples at the same time, each sample is repeated twice, a positive control, a negative control and a blank control. The detection time is only 100 minutes.
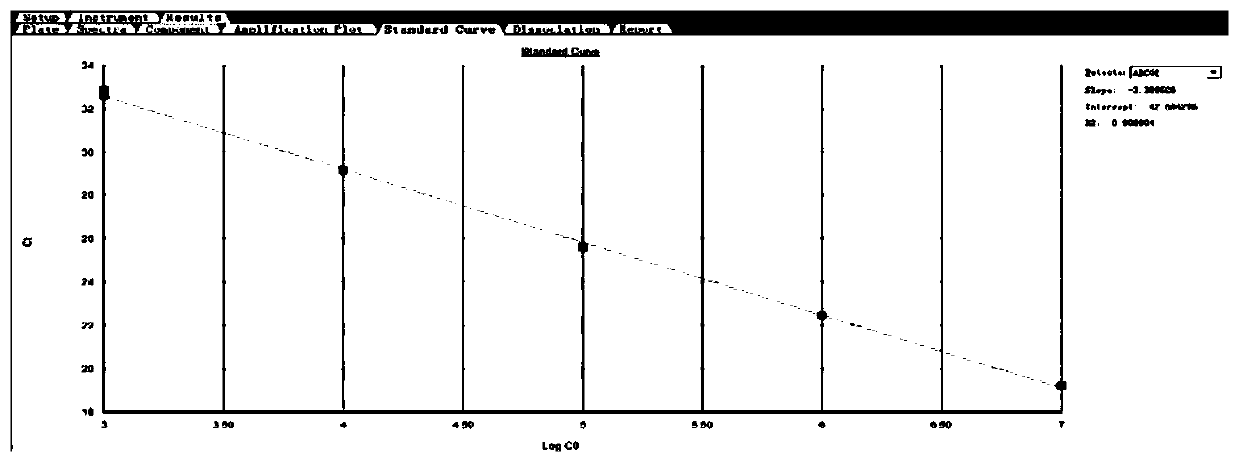
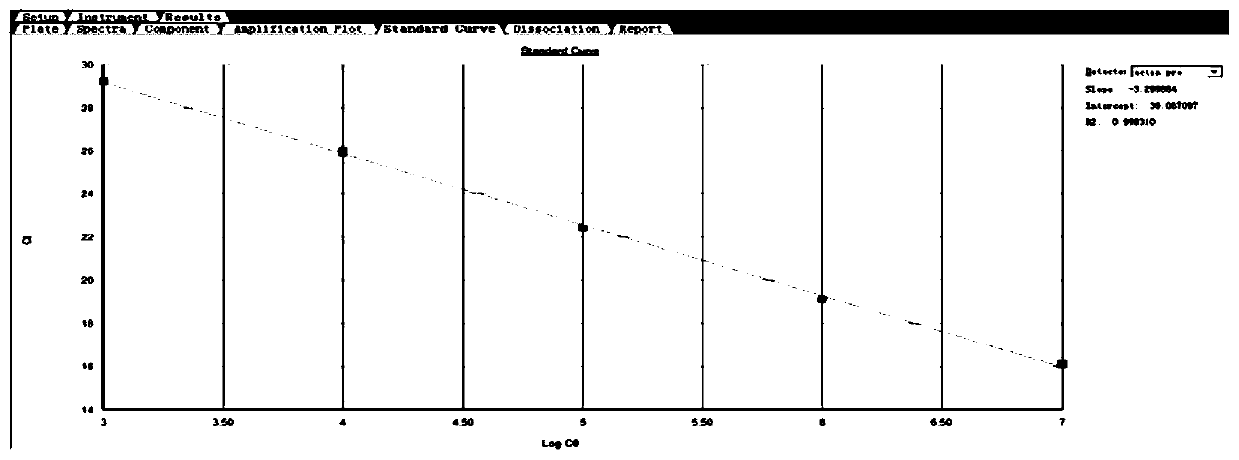
[0090] Test results such as Figure 5 As mentioned above, the normal human blood cDNA was used as a template to detect ABCG2 and...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com