A method for automatic analysis of non-targeted metabolic profile data in uplc-hrms Profile mode
A metabolic profile and automatic analysis technology, applied in the field of ultra-high performance liquid chromatography-high resolution mass spectrometry data analysis, can solve the problems of determination of interfering substances, redundant results, wrong judgments, etc. adaptable effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
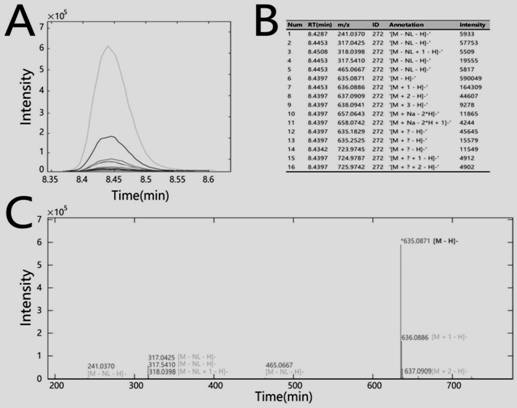
[0025] Below in conjunction with the accompanying drawings of the present invention, the technical solutions implemented by the invention will be further explained.
[0026] In the following, tea leaves and licorice were used as samples, and Agilent, Thermo and Waters instruments were used for UPLC-HRMS analysis.
[0027] S1: The preparation process of the tea samples analyzed by Agilent: the obtained Longjing tea samples were freeze-dried and crushed, and 1mg, 2mg, 3mg, 4mg, 5mg, 6mg of the crushed samples were weighed respectively, and 1.5mL of the extract was added to each sample. Said extract comprises methanol and water and 0.98mg / L internal standard umbelliferone in a volume ratio of 1:9, after vortexing for 2 minutes, ultrasonication at room temperature for 30 minutes, and then centrifugation at 13000r / s for 10min , transfer 1 mL of the supernatant to a chromatographic vial for analysis by UPLC-HRMS.
[0028] S2: Agilent 1290-6545 UPLC-QTOF analysis:
[0029] The chro...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com