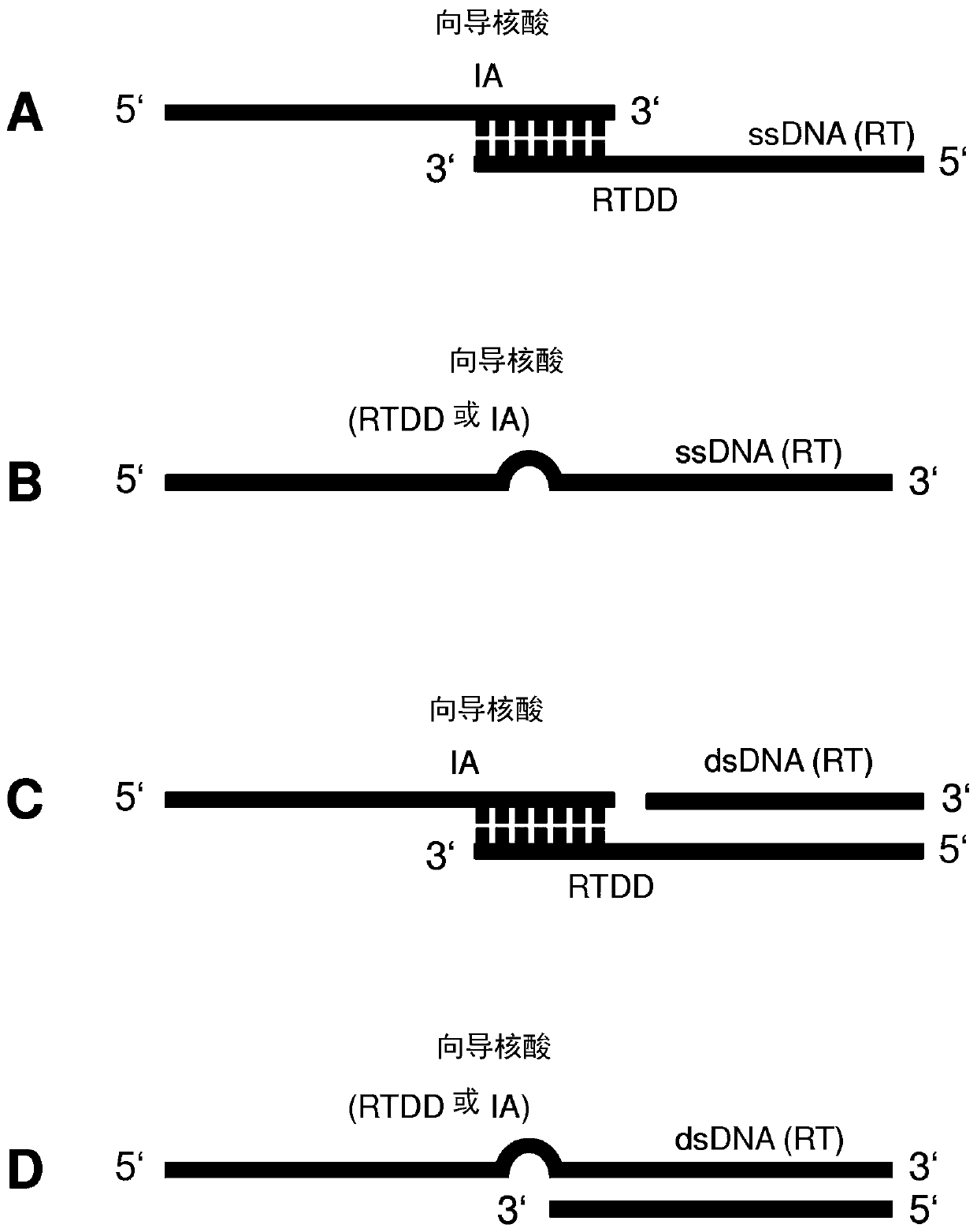
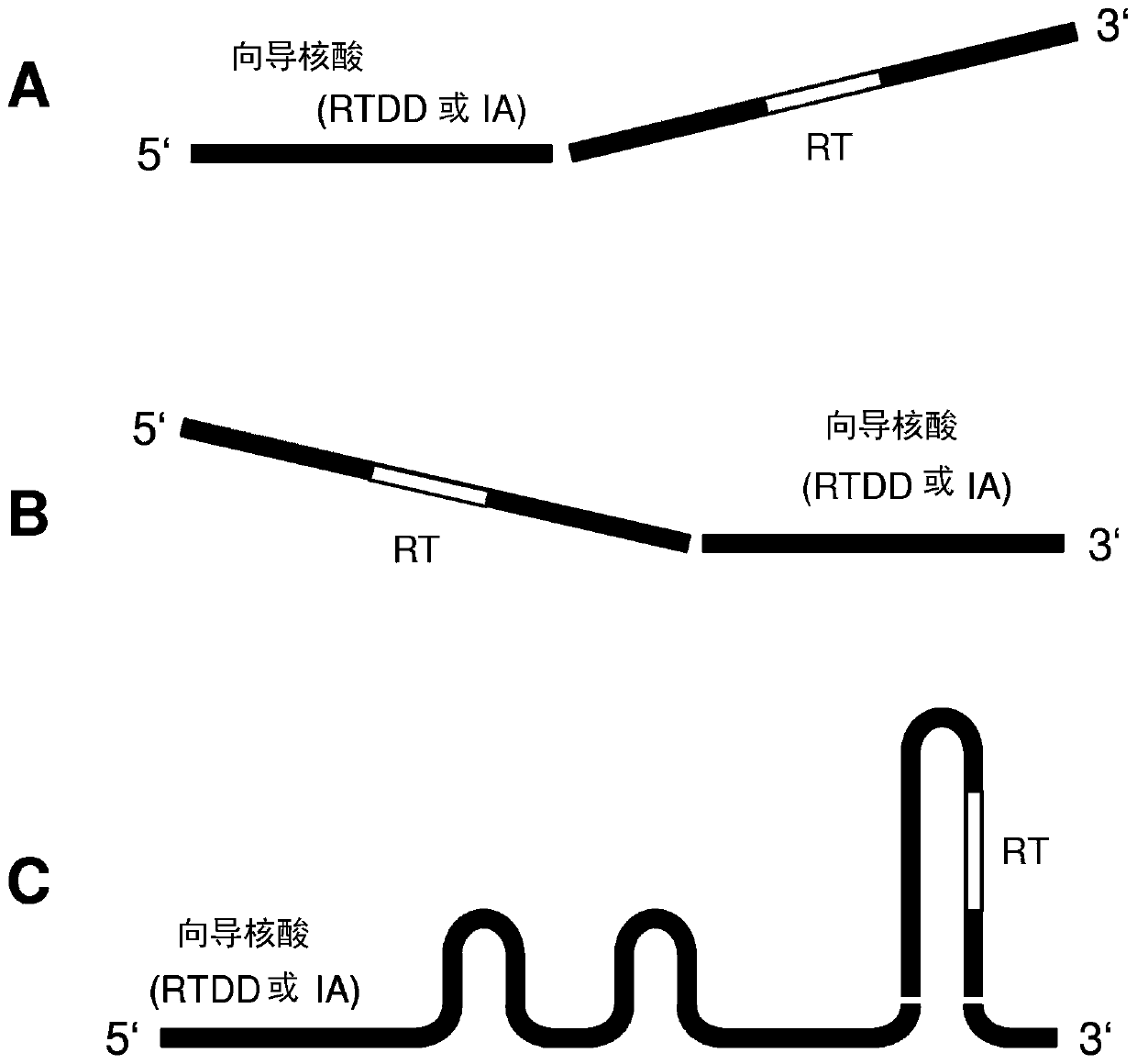
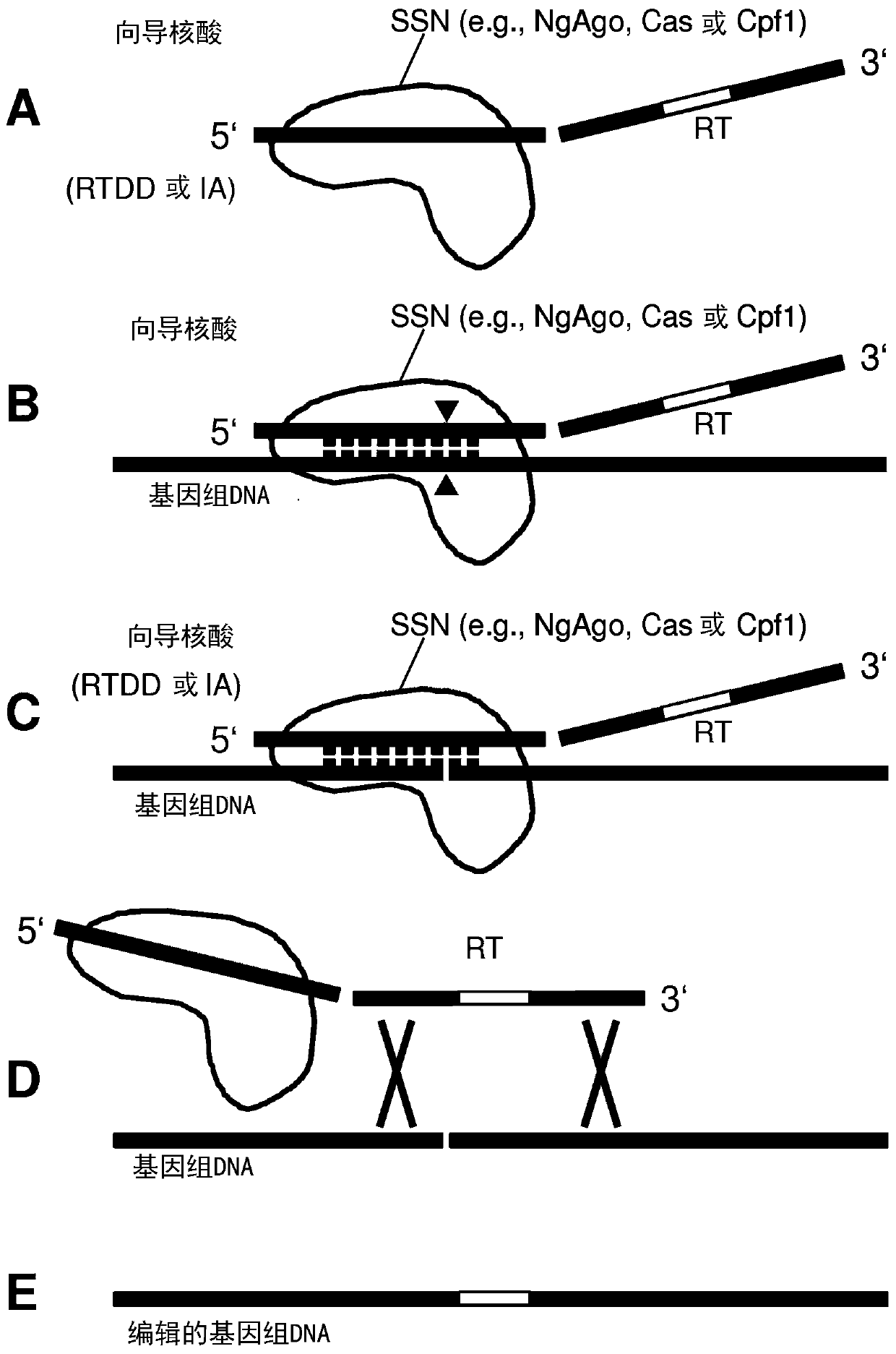
Repair template linkage to endonucleases for genome engineering
A nuclease and genome technology, used in genetic engineering, plant genetic improvement, applications, etc., can solve the problems of discovering and using site-specific nucleases and repair templates that have not yet been exploited, the process is error-prone, and the efficiency is low.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0289] Embodiment 1: be suitable for being combined with Cas or Cpf1 or Argonaute polypeptide as the hybrid nucleic acid sequence of RTDD / RT pair
[0290] In one experiment, tailed sgRNAs or sgDNAs were hybridized via complementary base pairing and RNA-DNA or DNA-DNA ligated single-strand repair templates. For covalent conjugation, synthetic DNA oligonucleotides are manipulated with the manufacturer's ssRNA ligase to covalently join the 3' ends of RNA / DNA oligonucleotides. For non-covalent conjugation, RNA / DNA and DNA oligonucleotides with partially complementary sequences were mixed and allowed to complex via Watson Crick base pairing. Successful hybridization can be determined in a gel shift assay. Treatment of hybrid nucleic acid aliquots with RNase and DNase enzymes prior to gel shift assays revealed that, for those experiments using sgRNA, some hybrid nucleic acids consisted of RNA and some consisted of DNA. The nucleic acid hybrid is then complexed with a recombinant C...
Embodiment 2
[0291] Example 2: In vitro cleavage of DNA targets by complexes of Cas9 protein and hybrid RNA-DNA nucleic acids
[0292] In one experiment, the Cas9 protein was tested for its function as a site-specific endonuclease when used with the described nucleic acid hybridization technology. A linearized plasmid containing at least one target site for the sgRNA is mixed with the Cas9-sgRNA-RT complex as described in the present invention. After incubation under conditions suitable for nuclease activity, including correct pH, temperature, and cofactors, etc. known to those skilled in the art for a variety of CRISPR nucleases and their variants, the DNA target plasmids are run on an agarose gel And observe the band size indicating the expected target site for cleavage. In vitro cleavage of target DNA demonstrated that RT associated with sgRNA as "cargo" did not interfere with the function of the Cas9 complex as a site-specific endonuclease.
Embodiment 3
[0293] Example 3: In vivo editing by Cas9 protein complexed with hybrid RNA-DNA nucleic acid
[0294] To demonstrate that target genes can be edited in vivo by delivering a complex containing the Cas9 protein and hybrid RNA-DNA nucleic acid, the non-functional tdTomato gene contained within the transformed plasmid was repaired by exchanging a single nucleotide to restore the fluorescent signal from the tdTomato gene . To determine optimal use for editing by providing ssDNA repair templates that are complementary to the target or non-target strand, complexes carrying either strand repair templates were compared.
[0295] The hybrid nucleic acid RNA / DNA-Cas polypeptide complex obtained in Example 1 was used to repair the episomal plasmid target encoding the tdTomato gene with a single point mutation from A to T at codon position 51 Generate an early termination signal. This plasmid was introduced into maize protoplasts by PEG- or electroporation-mediated delivery of an editing...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com