Methylation protected host bacterium and construction method and application thereof
A technology of host bacteria and methylation, applied in the biological field, can solve the problems of non-methylation modification, low expression level, narrow application range, etc., and achieve the effect of normal expression
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
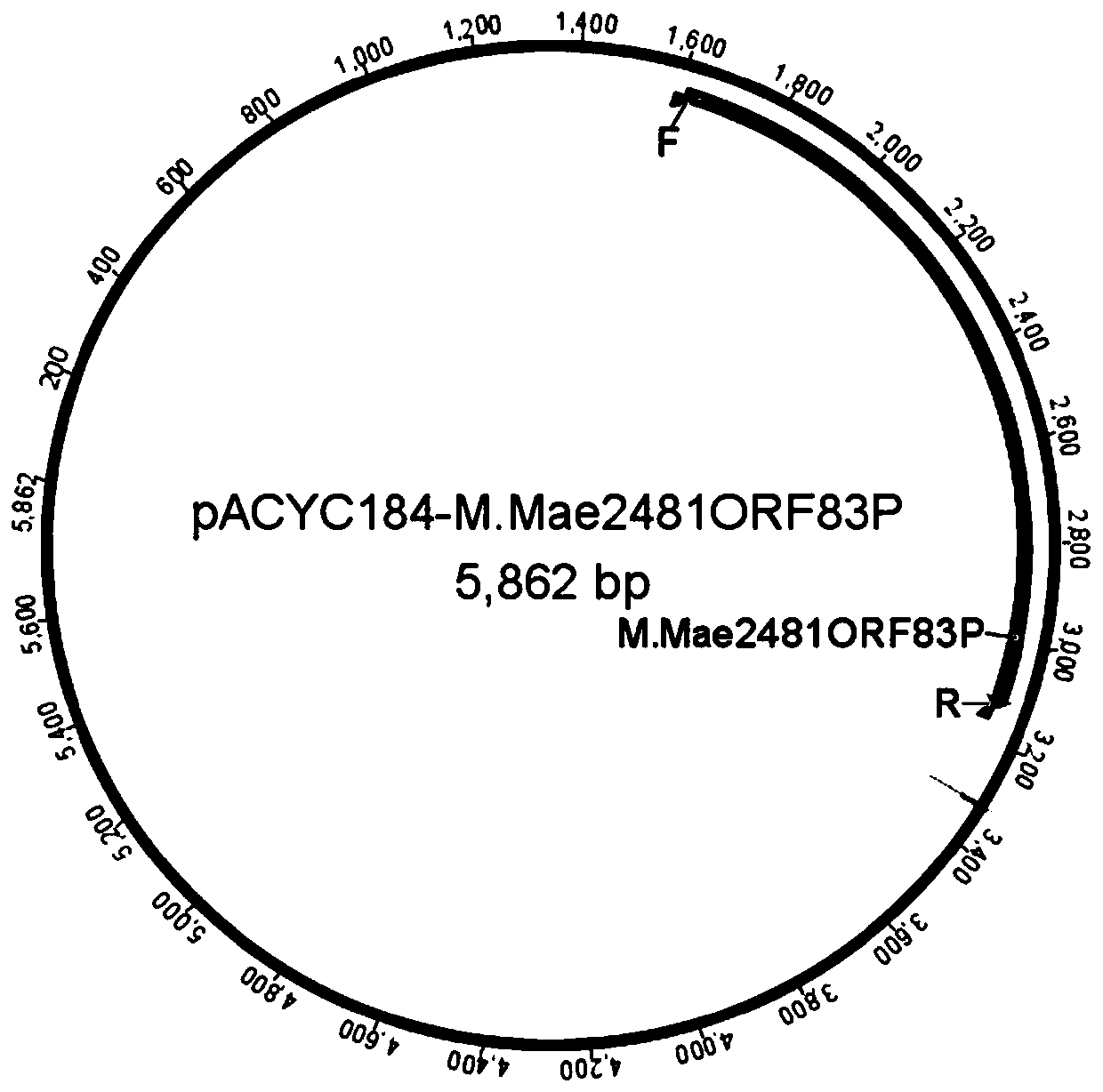
[0076] Example 1 Construction of Methyltransferase M.Mae2481ORF83P Host Bacteria
[0077] (1) Obtain the methyltransferase M.Mae2481ORF83P gene
[0078] The PCR amplification method was used to obtain the methyltransferase M.Mae2481ORF83P gene from the genome of the bacterial strain Microcystis aeruginosaNIES-2481 using specific primers for the methyltransferase M.Mae2481ORF83P gene. The specific steps were as follows:
[0079] Plate-culture colony of strain Microcystis aeruginosaNIES-2481 resuspended in deionized water, incubated at 95°C for 10min, used as PCR template, nucleic acid sequence of SEQ ID NO:3-4 as PCR primer pair, using MonHI-FI DNAPolymerase, according to the standard procedure The PCR reaction obtained the M.Mae2481ORF83P gene fragment which was consistent with the theoretical value.
[0080] (2) Construction of methyltransferase expression vector
[0081] pACYC184 plasmid using FlashCut TM NdeI and FlashCut TM After XhoI double digestion, with MonClone ...
Embodiment 2
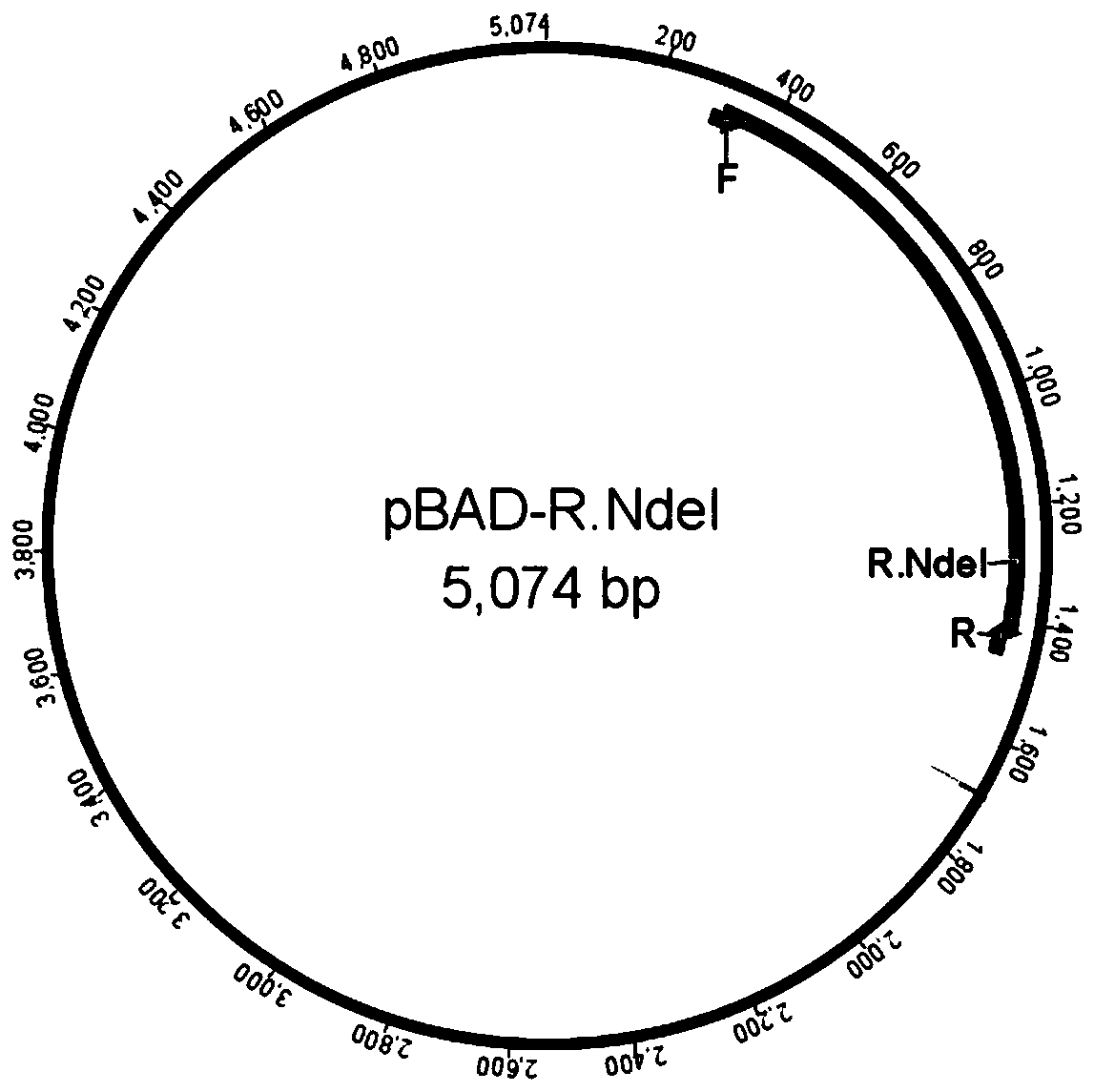
[0087] The construction of embodiment 2 restriction endonuclease NdeI recombinant expression strains
[0088] (1) Obtain restriction endonuclease NdeI gene
[0089] Using the method of PCR amplification, use the specific primer of restriction endonuclease NdeI gene from the genome of bacterial strain Neisseria denitrificans, obtain restriction endonuclease NdeI gene, specific steps are:
[0090] Plate-cultured colonies of the strain Neisseria denitrificans resuspended in deionized water, incubated at 95°C for 10 minutes, used as a PCR template, the nucleic acid sequence of SEQ ID NO:5-6 as a PCR primer pair, using MonHI-FI DNA Polymerase, and performing PCR according to the standard procedure The reaction obtained the R.NdeI gene fragment consistent with the theoretical value.
[0091] (2) Construction of restriction endonuclease NdeI expression vector
[0092] pBAD plasmid using FlashCut TM NdeI and FlashCut TM After HindIII double enzyme digestion, dephosphorylate with M...
Embodiment 3
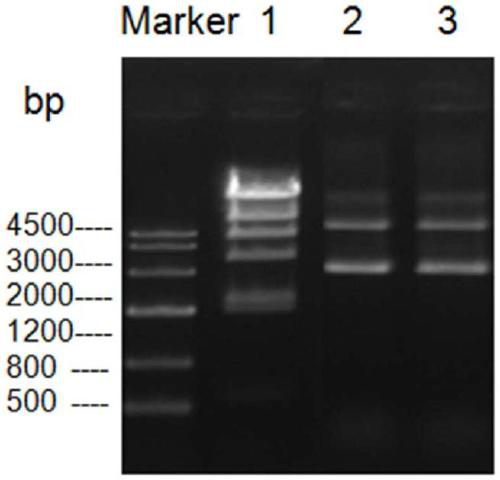
[0098] The activity detection of embodiment 3 restriction endonuclease NdeI
[0099] The enzymatic activity of restriction endonuclease NdeI is defined as: 1 μL of restriction endonuclease NdeI can completely digest 1 μg of λDNA within 15 minutes in a 20 μL reaction system at 37°C.
[0100] In this example, the purified restriction endonuclease NdeI was serially diluted in 1×FlashOne TM Buffer (Mona Biotechnology Co., Ltd.) buffer conditions, in a 20 μL reaction system, incubated with the substrate λDNA at 37° C. for 60 min, and then detected the digestion of the substrate by 1% agarose gel electrophoresis. The result is as Figure 4 As shown, the purified restriction enzyme NdeI has good enzyme cutting activity, and the specific activity is about 900,000 U / mg induced bacterial liquid.
[0101] In summary, the present invention transforms the M.Mae2481ORF83P methyltransferase expression vector into the host bacteria to construct a methylation-protected bacterial strain, whi...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com