Method for verifying feasibility of inserting CRISPR-Cas9 system mediated target gene into candida utilis
A technology for producing Candida utilis and target genes, which can be applied to other methods of inserting foreign genetic materials, microorganism-based methods, genetic engineering, etc., and can solve problems such as unidentified
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0097] CRISPR-Cas9-mediated targeted integration and expression of reporter gene GFP
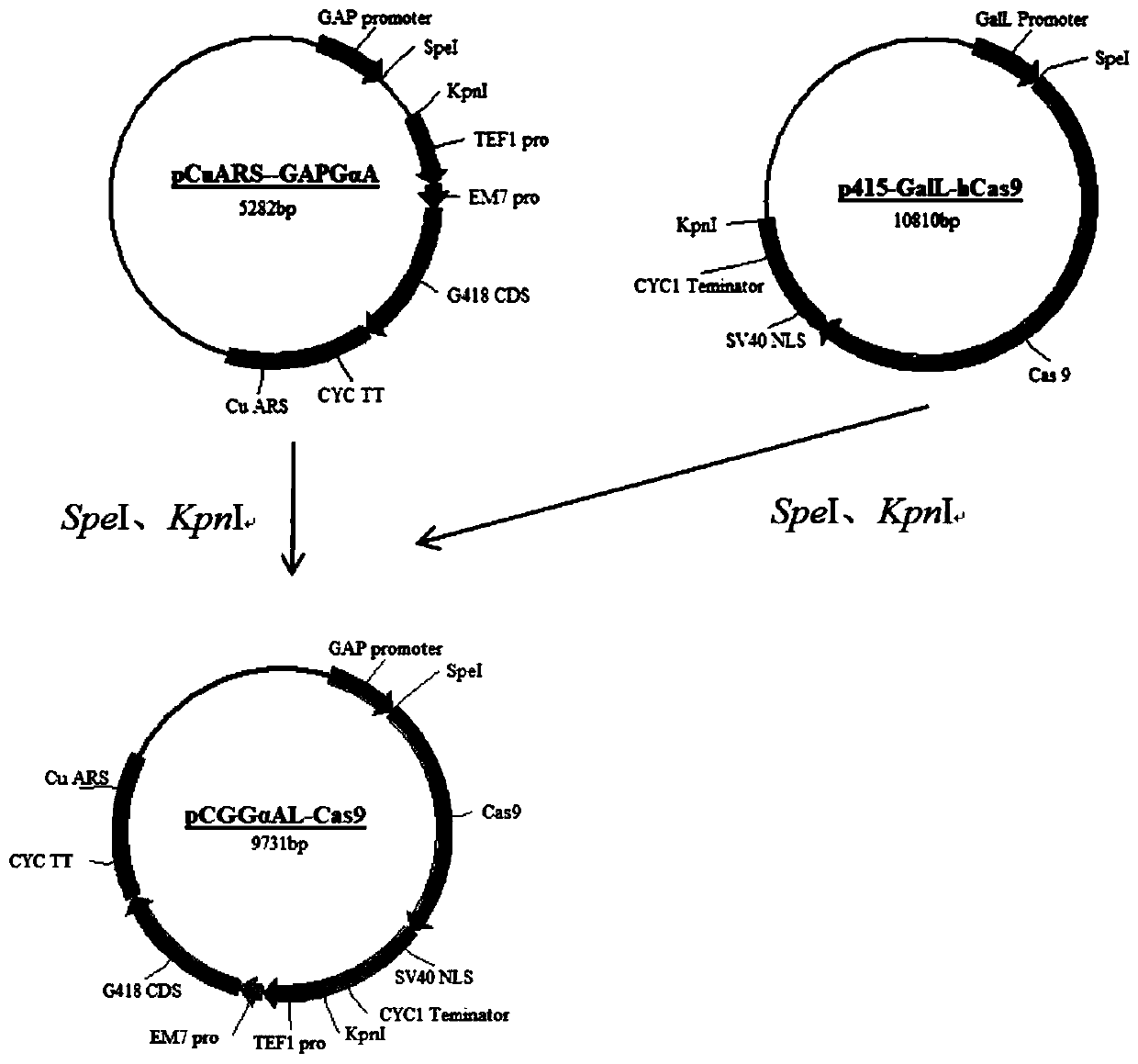
[0098] Candida utilis ATCC22023 used in this example was purchased from Guangdong Microbial Culture Collection Center; yeast Cas9 expression vector p415-GalL-hCas9 and sgRNA expression vector p426-crRNA were provided by Addgene; Candida utilis free expression Vector: pCuARS-GAPGαA, pCuARS-GAPZαAm, the structure and special sequence of pCuARS-GAPGαA are as follows Figure 24 As shown, the structure and special sequence of pCuARS-GAPZαAm are as Figure 25 Shown; Escherichia coli cloning vector pMD19-T was purchased from Dalian Bao Biological Engineering Co., Ltd.; primer synthesis and DNA sequencing were completed by Invitrogen; primer sequence-F represents the upstream primer, and primer sequence-R represents the downstream primer.
[0099] 1. Construction and verification of Candida utilis Cas9 expression vector
[0100] 1. Extract the plasmids p415-GalL-hCas9 and pCuARS-PGAPGαA, and then ...
Embodiment 2
[0236] CRISPR-Cas9-mediated targeted insertion of pectin methylesterase gene into the genome of Candida utilis
[0237] The expression vector used in this example is from the same source as the expression vector in Example 1 above. Primer synthesis and DNA sequencing were completed by Invitrogen Company. Candida utilis episomal expression vector: pCuARS-PGKG-GFP, its specific structure and special sequence such as Figure 26 Shown; Plasmid pCGGαAL-pmeA (containing the GAP promoter of Candida utilis, G418 resistance gene) with pmeA gene, its specific structure and special sequence are as follows Figure 27 shown. Both the pGlu-sgRNA-PGK plasmid and the Candida utilis containing Cas9 were constructed in Example 1; the primer sequence-F represents the upstream primer, and the primer sequence-R represents the downstream primer.
[0238] Based on the existing pGlu-sgRNA-PGK plasmid and Cas9-containing Candida utilis, a recombinant donor fragment containing left and right homology...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com