SSR core primer group developed based on whole genome data of tartary buckwheat and application thereof
A whole-genome, core primer technology, applied in the field of molecular biology, can solve the problems that cannot meet the needs of tartary buckwheat research, and the number of tartary buckwheat SSR markers is small
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
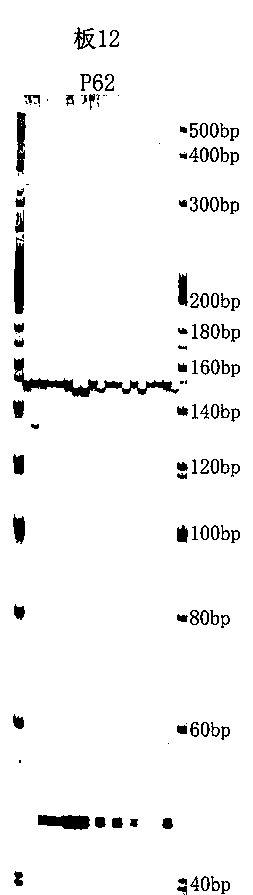
[0029] High-throughput acquisition of SSR markers in tartary buckwheat genome
[0030] SSR marker search and primer design
[0031] 1. Obtain the tartary buckwheat genome sequence
[0032] The tartary buckwheat genome sequence completed by the research group was used as the template sequence.
[0033] 2. Search for genome sequences containing SSRs
[0034] Install the perl language, download the misa.pl program from http: / / pgrc.ipk-gatersleben.de / misa / misa.html, name the sequence file containing Unigene as S.fasta, and copy it to the perl\bin directory of the C drive, in Execute the command C:\perl\bin\perlmisa.plS.fasta in the perl environment, search for the SSR site in the sequence and get two files, S.fasta.misa and S.fasta.statistics. Among them, the minimum repetition numbers of 1-6 bases are 10, 6, 5, 5, 5, and 5 respectively, and the distance between two SSRs is less than 100bp, and they are merged into a composite SSR.
[0035] Table 2 SSR length statistics table ...
Embodiment 2
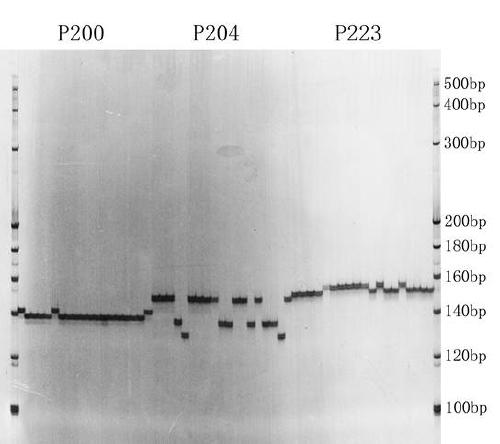
[0049] Example 2 Polymorphism Verification of SSR Primers
[0050] The 5 tartary buckwheat resources and 3 sweet buckwheat resources recorded in Table 3 with large phenotype differences were used for planting, and 2 g of leaves at the seedling stage were collected respectively, and the leaves were crushed with a high-throughput pulverizer, and Shanghai Sangong DNA extraction kit was used. (Ezup Column Plant Genomic DNA Extraction Kit, B518261-0050) for leaf DNA extraction, DNA quality detection and concentration determination on a microplate reader, and DNA was diluted to a final concentration of 30ng / μL, and kept at -20°C save. The polymorphisms of the primers were verified by PCR amplification using the material DNA and the randomly selected pair of SSR primers. The PCR reaction system was 10 μL in total, including 1.5 μL (30ng / μL) of genomic DNA, 5 μL of 2×TaqPCR MasterMix (Tiangen, KT: 121221), and 2 μL (0.002 nmol / μL) of forward and reverse primers in total. The PCR amp...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com