CPA primers and kit for detection of methicillin-resistant staphylococcus aureus, and detection method
A methicillin-resistant Staphylococcus aureus and methicillin-resistant technology, applied in the field of biotechnology detection, can solve problems such as insufficient sensitivity, limited scope of application, etc., and achieve the effects of simple and fast operation, good applicability, and shortened cycle
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0045] The method for detecting methicillin-resistant Staphylococcus aureus based on cross-primer constant temperature amplification (CPA) reaction technology may further comprise the steps:
[0046] (1) Reagents used:
[0047] a. The stripping primers 4s and 5a, the cross-amplification primers 2a1s, and the specific primers 2a and 3a each at a concentration of 10 μM, the primer sequences are as shown in the preceding SEQ ID NO.1-SEQ ID NO.10;
[0048] b.2×Reaction stock solution: Tris-HCl with concentration of 40.0mM, ammonium sulfate of 20.0mM, potassium chloride of 20.0mM, magnesium sulfate of 16.0mM, Tween 20 of 0.2% (v / v), 1.4M Betaine, 10.0mM dNTPs (each) composition;
[0049] c. Bst DNA polymerase (large fragment, NEB company) aqueous solution with a concentration of 8U / μL;
[0050] d. Mixed solution of calcein and manganese chloride: first prepare a calcein solution with a concentration of 50 μM (dissolved in dimethyl sulfoxide); then take 25 μL of 50 μM calcein solu...
Embodiment 2
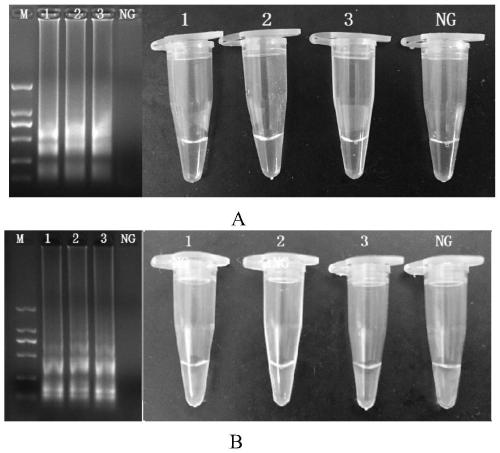
[0060] Cross constant temperature amplification reaction (CPA) detection methicillin-resistant Staphylococcus aureus specific test, comprises the following steps:
[0061] The genomic DNA of methicillin-resistant Staphylococcus aureus NCTC10442 and non-methicillin-resistant Staphylococcus aureus was established according to the reaction system and conditions in Example 1 to establish a cross-isothermal amplification reaction detection method, and a specificity test was carried out;
[0062] Among them, the non-methicillin-resistant Staphylococcus aureus is: Escherichia coli O157:H7E019; Escherichia coli O157:H7E020; Escherichia coli E043; Escherichia coli E044; Listeria ATCC19116; Listeria monocytogenes ATCC19114; Listeria monocytogenes ATCC19115; Listeria monocytogenes ATCC15313; Listeria monocytogenes ATCC19113; Pseudomonas aeruginosa ATCC27853; Vibrio parahaemolyticus ATCC17802; ATCC27969; Lactobacillus casei BM-LC14617.
[0063] The genome of methicillin-resistant Staphyl...
Embodiment 3
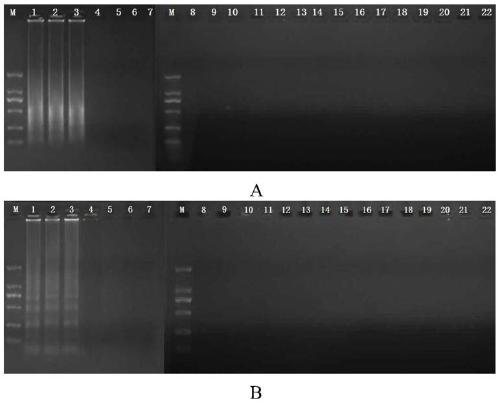
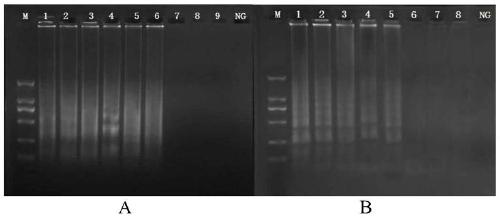
[0065] Cross constant temperature amplification reaction (CPA) detects the sensitivity comparison test of methicillin-resistant Staphylococcus aureus, comprising the following steps:
[0066] The genome of methicillin-resistant Staphylococcus aureus NCTC10442 was serially diluted 10 times to 3.0ng / μL, 300pg / μL, 30pg / μL, 3pg / μL, 300fg / μL, 30fg / μL, 3fg / μL, 300ag / μ L, set negative control (nucleic acid-free water) simultaneously, construct the cross constant temperature amplification method according to the reaction system among the embodiment 1 and carry out 2% agarose gel electrophoresis to the amplified product, to determine the sensitivity of detection method.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com