Application of crRNA targeted PCR-CRISPR system to detection of HBVDNA
A system and purpose technology, applied in the direction of recombinant DNA technology, DNA/RNA fragment, microbial determination/inspection, etc., can solve the problems of high cost, prone to contamination, not suitable for clinical detection, etc., to achieve simple method, high sensitivity and specific effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0040] Embodiment 1: be used for the design and preparation of crRNA of the present invention and PCR primer
[0041] (1) Design and preparation of crRNA
[0042] The detection target of HBV DNA is selected near the P area of hepatitis B virus. First, the conservative analysis of this part of the sequence was carried out, and the hepatitis B virus database ( https: / / hbvdb.ibcp.fr ) to download the sequence information of the P region of 7720 strains of hepatitis B virus, apply clustal X software to carry out bioinformatics analysis on the sequence, and use the Perl script to analyze the base ratio of each site. Then select and design corresponding PCR amplification primers and crRNA according to the analysis results.
[0043] The present invention selects the conserved sequence of the reverse transcriptase region (RT region) to design crRNA. The 5' end of the crRNA has a 39nt repeat sequence, which can bind to LwCas13a protein, 5'-GGGGAUUUAGACUACCCCAAAAACGAAGGGGACUAAAAC-...
Embodiment 2
[0068] Example 2. Establishment of the method for detecting HBV DNA by CRISPR-Cas13a
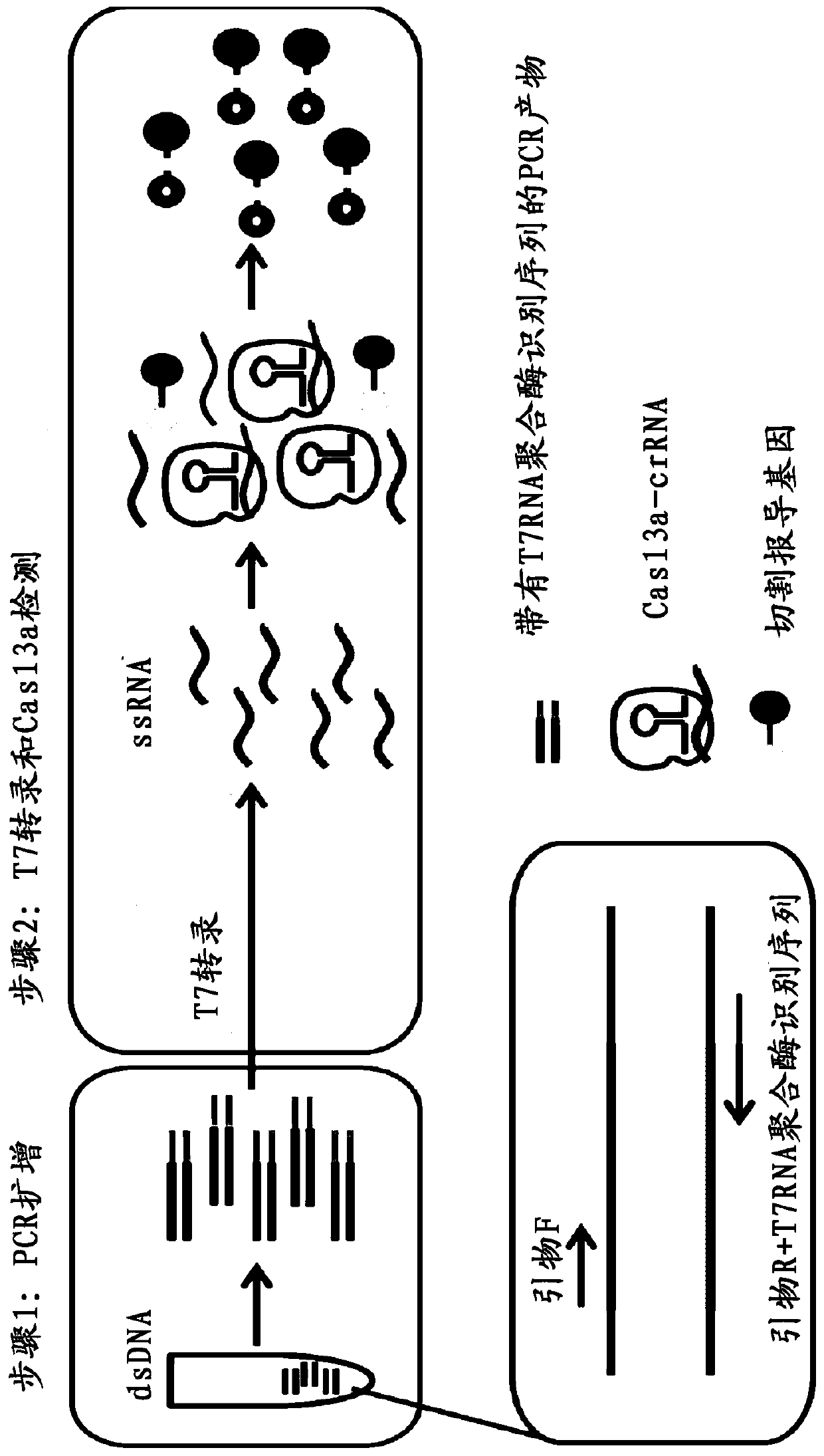
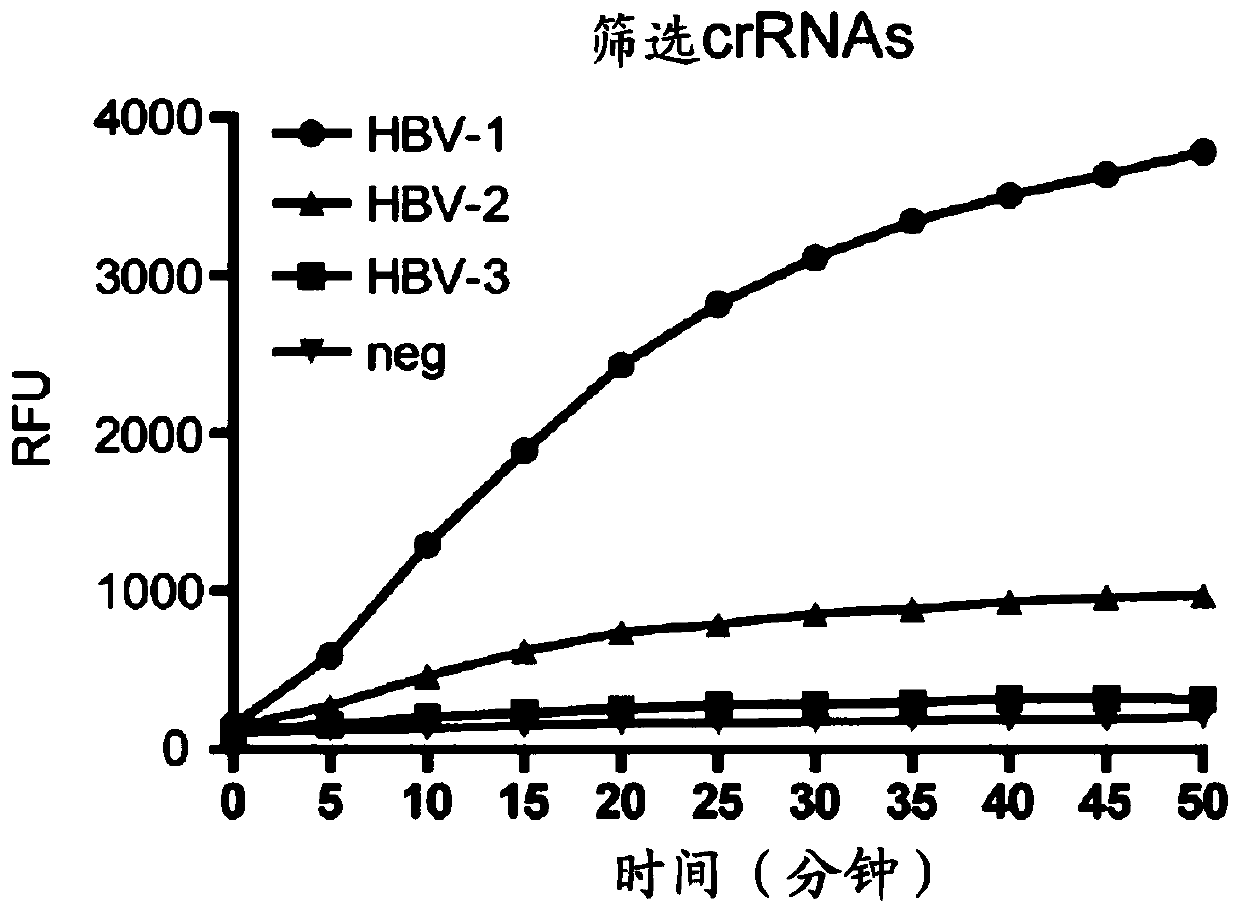
[0069] In this study, PCR was used to amplify the target nucleic acid, and the target sequence detected by HBV DNA was transcribed to obtain the corresponding ssRNA, and the above crRNAs: HBV-crRNA1, HBV-crRNA2, HBV-crRNA3 were used for detection, and the results of different crRNAs were compared. Signal intensity, select the crRNA with the strongest fluorescent signal as the subsequent crRNA detection. Specific steps and principles such as figure 1 Shown: the first step uses specific primers to amplify the target sequence (through denaturation, annealing, and extension processes), and there is a T7 transcription sequence at the 5' end of the primer, so that the double-stranded DNA obtained by PCR amplification ( dsDNA) can be recognized and transcribed by T7 RNA polymerase. In the second step, part of the amplified product was taken out and added to T7 RNA polymerase, LwCas13a protein, cr...
Embodiment 3
[0079] Example 3. Sensitivity of CRISPR-Cas13a to detect HBV DNA
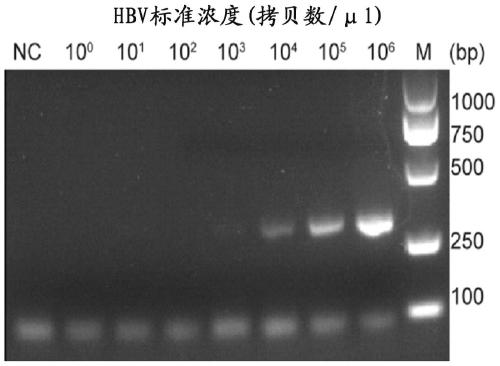
[0080] The sensitivity experiment mainly detects the lowest copy number of hepatitis B virus that can be detected by this method, and compares it with the existing fluorescent quantitative PCR detection method. The plasmid pHBV1.2 was serially diluted to 10 6 、10 5 、10 4 、10 3 、10 2 、10 1 、10 0 Copies / ul, after PCR amplification, take 5ul of the amplified product and add it to the LwCas13a detection system, and set the PCR product with water as the template as a negative control. And take 2ul PCR products for agarose gel electrophoresis detection. At the same time, the hepatitis B virus DNA detection kit (Hunan Shengxiang Biotechnology Co., Ltd.) was used to detect different concentrations of plasmid templates, and the sensitivity of the two detection methods was compared. The agarose gel results of PCR amplification products are shown in image 3 , image 3 There is no band in the negative control (N...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com