S-adenosylmethionine synthetase mutant and high-throughput screening method thereof
A technology of adenosylmethionine and mutants, applied in botany equipment and methods, microorganism-based methods, biochemical equipment and methods, etc., can solve the problem of low catalytic activity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0024] Embodiment 1: Construction of eMAT recombinant bacteria
[0025] Select the MAT derived from Escherichia coli K12, use PCR technology, and use the E.coli K12 genome as a template to
[0026] metK-F:5'-GATCC GAATTC ATGGCAAAACACCTTTTTACG-3' (SEQ ID NO.3) and metK-R:5'-GTG CTCGAG TTACTTCAGACCGGCAGCAT (SEQ ID NO.4) was used as a primer to amplify the metK gene, and EcoR I and Xho I restriction endonuclease sites (underlined) were introduced at its 5' end and 3' end respectively. The PCR reaction system (50 μL) was: 25 μL of 2×PrimeSTAR Max Premix, 1 μL of template DNA, 1 μL of upstream and downstream primers, and 22 μL of sterile water. The PCR reaction conditions are: 98°C for 5 min; 98°C for 10s, 60°C for 5s, 72°C for 10s, cycle 30 times; 72°C for 5min. Verify the PCR amplification product with 1% agarose gel electrophoresis, when the target band size meets the size of the coding SEQ ID NO.1 ( figure 1 ), use PCR product recovery reagents and perform product recover...
Embodiment 2
[0027] Example 2: Establishment of eMAT catalytic activity transformation high-throughput screening method
[0028] Under acidic conditions, Pi can react with ammonium molybdate to form a blue complex. Malachite green can increase the color reaction of the complex, so that it has an obvious absorption peak at 620nm. Therefore, this chromogenic method is considered as a method for high-throughput screening. In order to ensure the sensitivity and accuracy of the screening process, the factors that may affect the color development in the experiment should be optimized first.
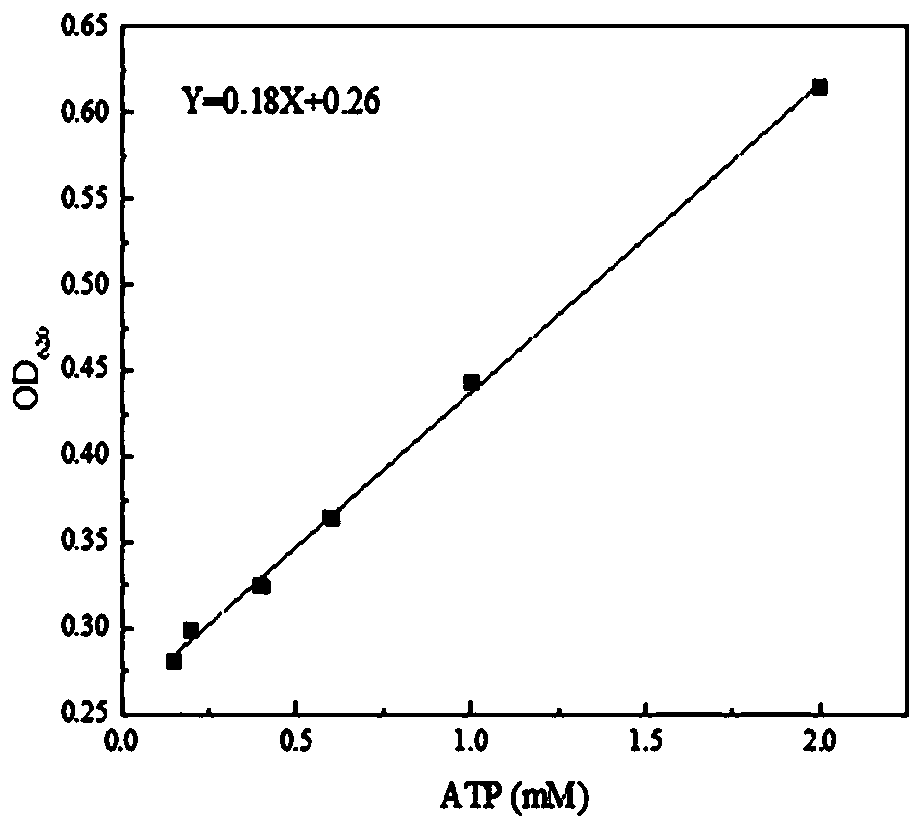
[0029] Using 100mM Tris-HCl as a control for color development, it was found that ATP would affect the color development ( figure 2 ). When the ATP concentration is lower than 2mM, OD 620 Increases linearly with increasing ATP concentration. But with the standard curve of Pi ( image 3 ), its slope is only 1 / 57. According to the fitting curve Y=0.18X+0.26, when ATP changes by 0.06mM, OD 620 The var...
Embodiment 3
[0033] Example 3: Construction and high-throughput screening of mutant libraries
[0034] Using the eMAT coding gene as a template, at 0.05mM, 0.04mM, 0.2mM Mn 2+ Error-prone PCR library construction was carried out under the following conditions, and 10 single colonies were randomly selected for sequencing, and it was found that only when Mn 2+ When the concentration is 0.04mM, the base mutation rate meets the requirements for error-prone PCR library construction (Table 2). The mutant library was initially screened by a high-throughput screening method, and 8 ODs were obtained 620 mutants at least 0.2 higher than controls. The 8 mutants were re-screened, and finally a mutant (R74H) with significantly higher specific enzyme activity than wild-type eMAT was obtained.
[0035] Table 2 Mn in PCR system 2+ The relationship between concentration and mutation rate
[0036]
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com