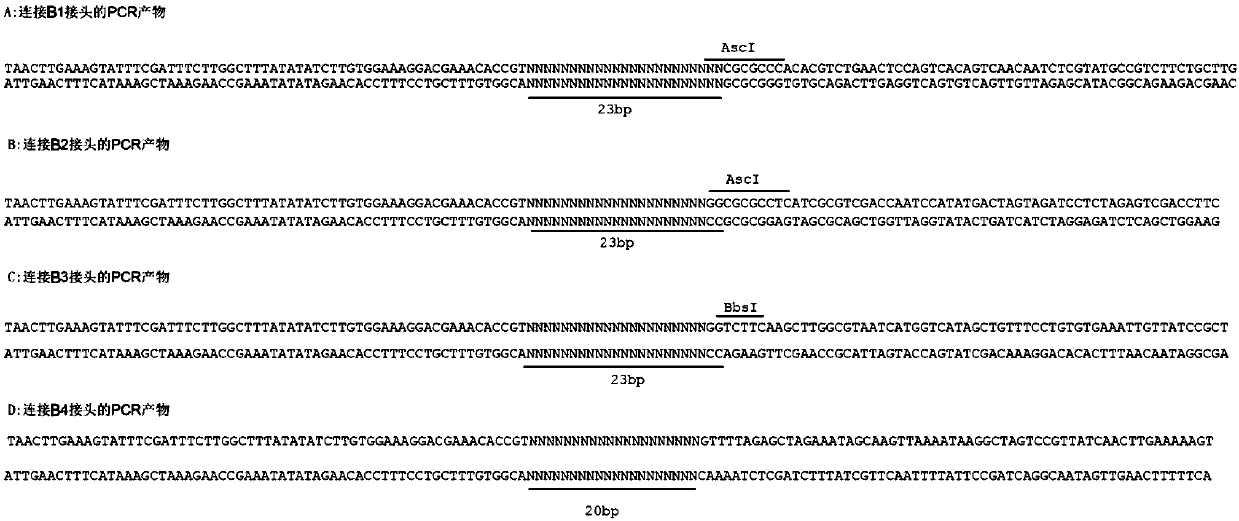
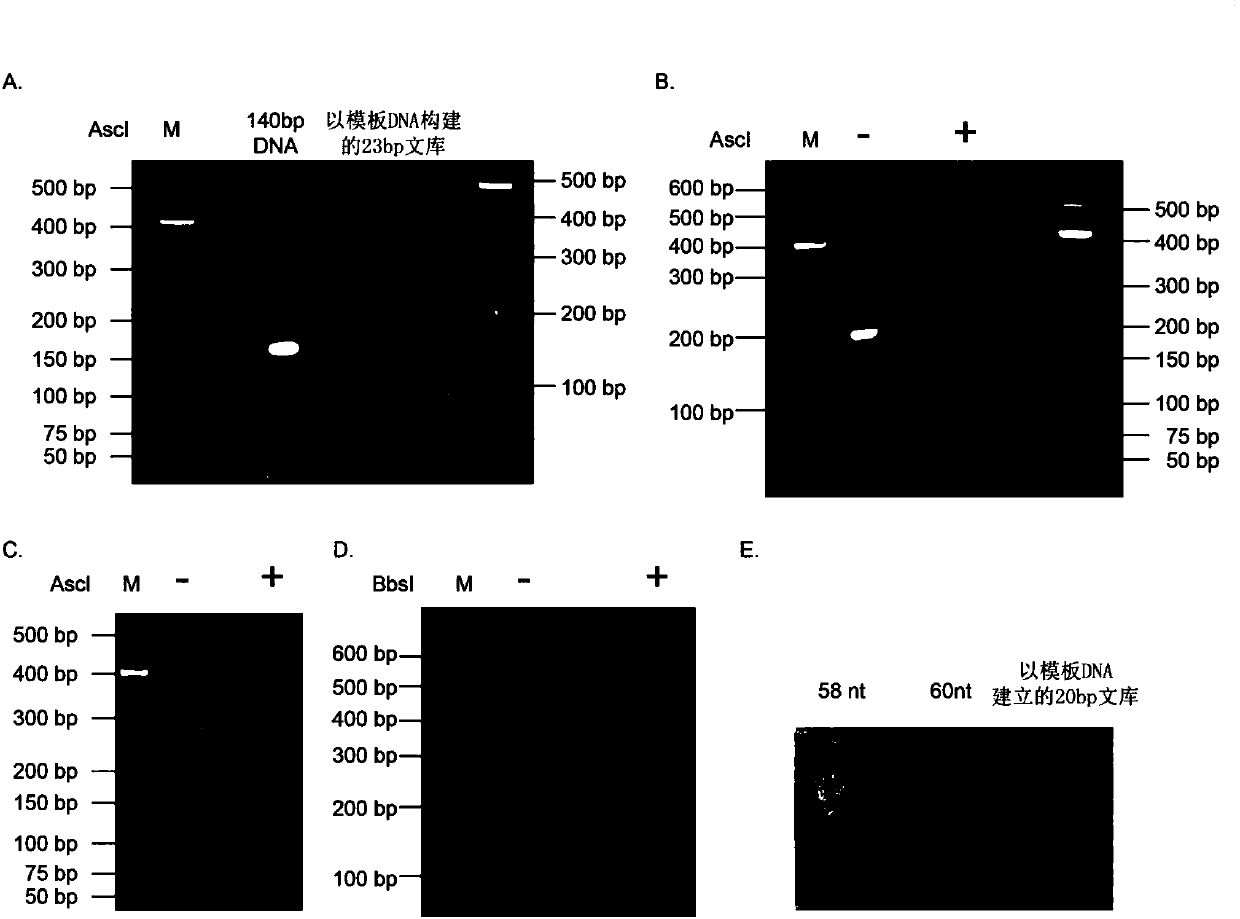
Method used for synthesis of DNA library with fixed length and specific terminal sequence based on template material
A DNA library, terminal sequence technology, applied in the field of synthetic biology
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0083] 1. Example 1 Starting material: Mouse embryonic stem cell H3K4me3 chromatin immunoprecipitation DNA
Embodiment 2
[0084] 2. Example 2 Starting material: Human embryonic stem cell H3K4me3 chromatin immunoprecipitation DNA
[0085] 3.NEBNext ultra-fast end repair / add dA tail module, NEB, E7442
[0086] 4. NEBNext ultra-fast connection module, NEB, E7445;
[0087] 5. Ampure XP beads, Beckman, A63881;
[0088] 6.Dynabeads TM T1 magnetic beads, Invitech, 65602;
[0089] 7.2X binding and washing buffer;
[0090] 8. Nuclease-free water, Ambion,10977023;
[0091] 9.1M sodium hydroxide, Sigma, S2770-100ML;
[0092] 10. Hybridization buffer;
[0093] 11. Extension buffer;
[0094] 12.3' hydroxyl recovery buffer;
[0095] 13. Washing buffer 1;
[0096] 14. Washing buffer 2;
[0097] 15.10X MBN buffer, NEB, M0250;
[0098] 16.MBN (10U / μL), NEB, M0250;
[0099] 17.10X T4DNA ligation buffer, NEB, B0202;
[0100] 18. T4 polynucleotide kinase, NEB, M0201;
[0101] 19. T4 DNA ligase, NEB, M0202;
[0102] 20. Qiaquick glue recovery kit, Qiagen, 28704;
[0103] 21. Qiaquick PCR product recovery kit, Qiagen, 28104;
[0104] 22.10X...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com