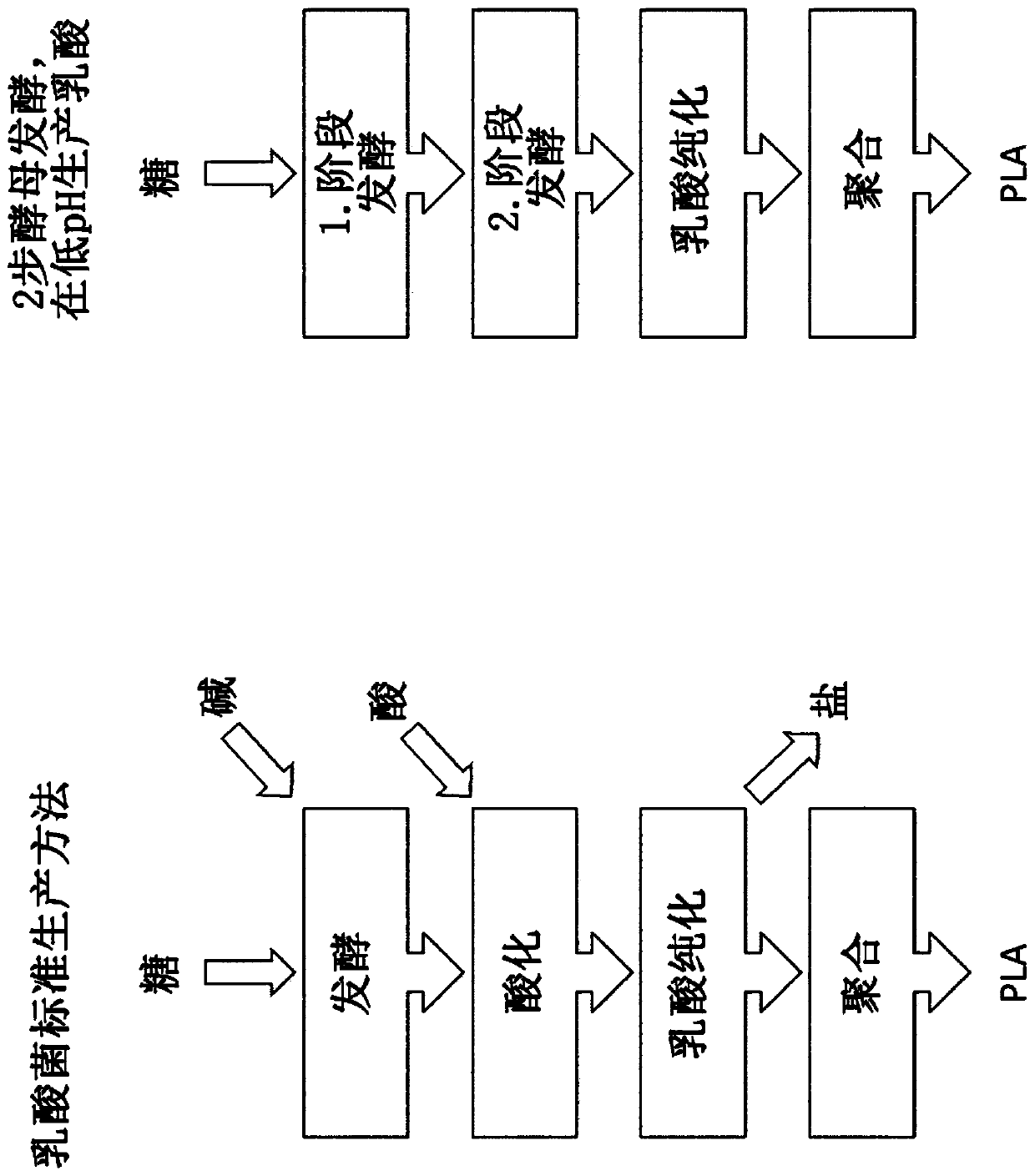
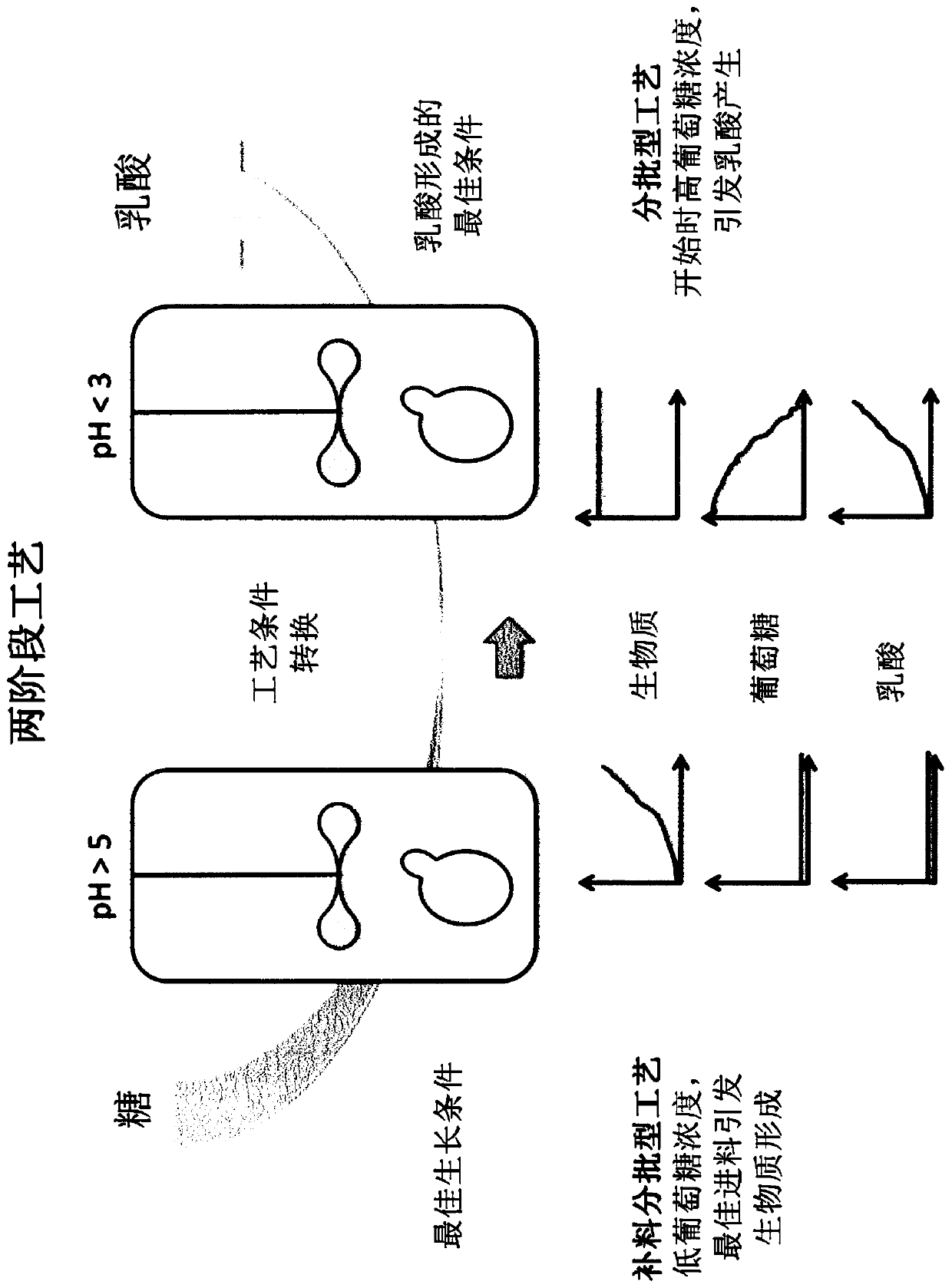
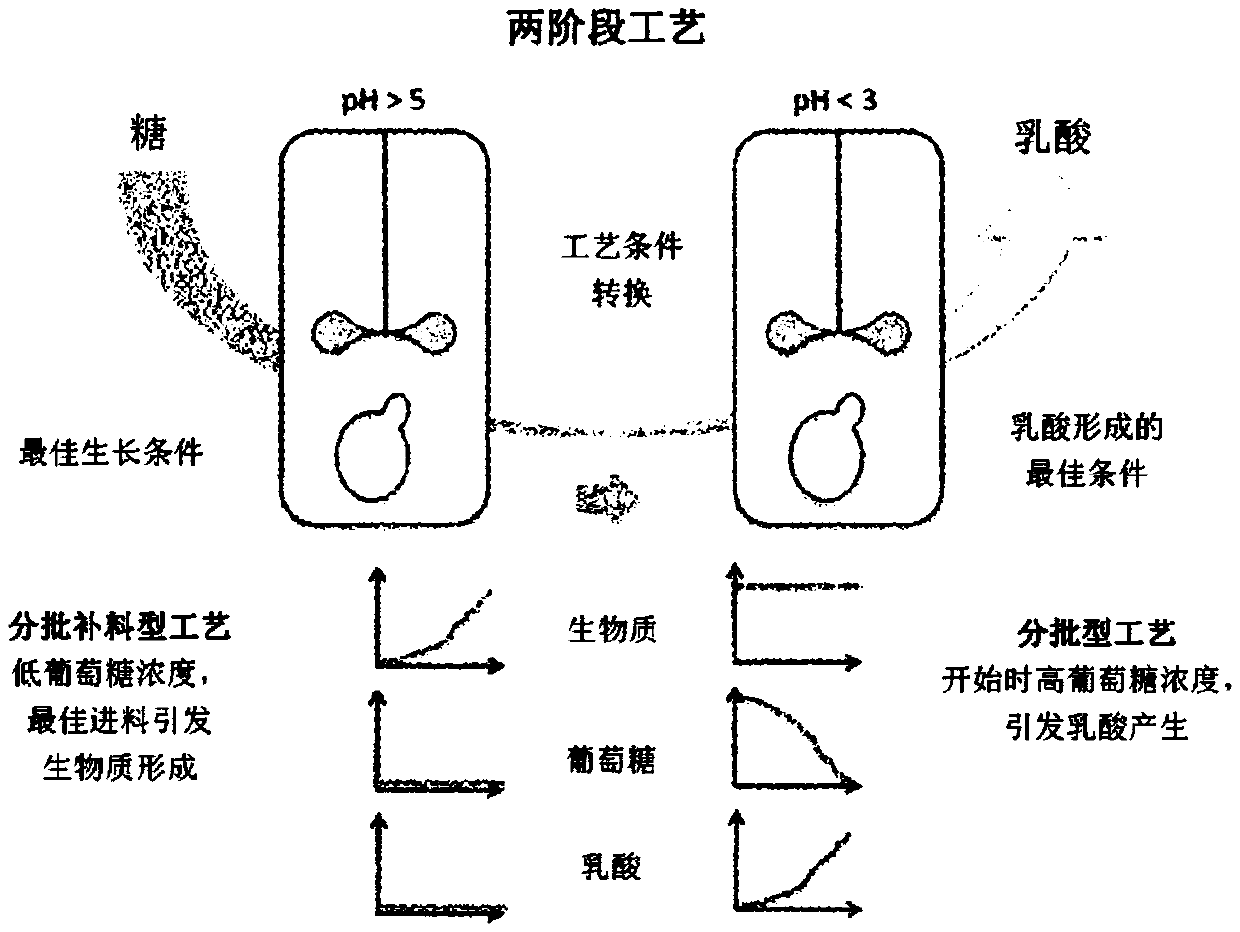
Method for producing lactic acid
一种乳酸、生物质的技术,应用在生物化学设备和方法、裂解酶、肽等方向,能够解决未满足改进工艺等问题,达到高产量、高生产率、降低生产成本的效果
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0257] Example 1: Characterization of the fermentation performance of Saccharomyces cerevisiae (CBS7962) strains at different glucose and lactic acid concentrations and pH.
[0258] The S. cerevisiae CBS7962 strain was grown in two stages in bioreactors and shake flasks. Use containing 1.8g / lYNB, 5g / l (NH 4 ) 2 SO 4 and 20g / l glucose medium for the inoculum incubation stage. Cells stored in glycerol vials stored at -80°C were used to inoculate 50 ml of preinoculum medium with an optical density at 600 nm (OD600) of 0.05. After 24 hours of growth, cells were harvested and used as inoculum during the culture phase in shake flasks and in bioreactors (Eppendorf DASGIPDASbox4, Germany) with working volumes of 50 ml and 700 ml, respectively. Table 1 and Table 2 list the medium composition and fermentation parameters of the culture stage. All cultures were carried out at 30°C; the shake flasks were incubated in an orbital shaker set at 180 rpm; potassium hydrogen phthalate buffe...
Embodiment 2
[0263] Example 2: Construction of S. cerevisiae strains lacking pyruvate decarboxylase (PDC1 , PDC5 and PDC6) genes using a split-maker deletion method.
[0264] Allelic knockout of PDC1, PDC5, and PDC6 was facilitated by a split marker deletion method using kanamycin and hygromycin as selection markers. The primer sequences used for deletion and validation primers are listed in Table 3. Two constructs were designed for deletion of one of the alleles. Each construct contained flanks of the target gene, followed by LoxP sites, and about half of one of the selectable markers. Flanking regions and LoxP sites were included in primers used to amplify part of the selectable markers for kanamycin and hygromycin from pPM2aK21 and pPM2a_hphR, respectively. Two PCR constructs containing target gene flanking regions, a LoxP site and part of one of the selectable markers were transformed into the CBS7962 strain. Transformation was performed according to the LiAc / PEG / ss-DNA protocol (ht...
Embodiment 3
[0267] Example 3: Construction of Saccharomyces cerevisiae strains with inactive pyruvate decarboxylase (PDC1, PDC5 and PDC6) genes: The pdc gene was knocked out using the CRISPR-cas9 system.
[0268] The CRISPR-cas9 system, which uses RNA-guided endonuclease activity to induce double-strand DNA breaks (dsbs) at target sites and repairs dsbs with double-strand (ds) DNA homologous oligonucleotides containing stop codons, is used to inactivate pdc gene. Transformation was performed according to the LiAc / PEG / ss-DNA protocol (http: / / home.cc.umanitoba.ca / ~gietz / method.html). Using golden gate assembly, the endonuclease cas9 was constructed in a centromeric plasmid under the TEF1 promoter and CYC1 terminator (Engler et al., 2008). Guide RNAs (gRNAs) for the PDC1 , PDC5 and PDC6 genes were identified using ChopChop (https: / / chopchop.rc.fas.harvard.edu / ). A 20 bp gRNA was constructed to contain the sequence of the hammerhead (HH) ribozyme and the sequence of the hepatitis D virus (H...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com