Intron 3-based Molecular Marker for Identifying Barley Semi-dwarf Multi-tiller Gene fol-a and Its Application
A fol-a, molecular marker technology, applied in the field of agricultural biology, can solve problems such as environmental pollution, increased fertilizer input, and increased production costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
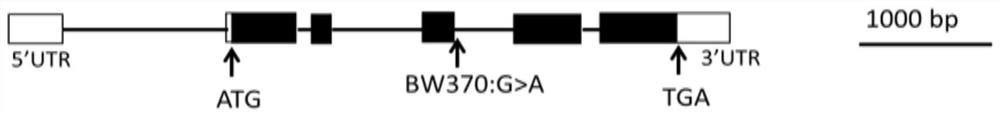
[0041] Embodiment 1, design molecular marker SNPE3
[0042]1), test materials: mutants BW370 and Bowman;
[0043] The gene fol-a controlling semi-dwarf and multi-tiller traits was cloned from the mutant BW370, and the nucleotide sequence of the gene is as described in SEQ ID NO:1. The protein encoded by the semi-dwarf multi-tiller gene fol-a has the amino acid sequence described in SEQ ID NO:3.
[0044] The radiation-induced mutant Proctor (the mutant has semi-dwarf and multi-tiller traits) was backcrossed with barley variety Bowman multiple times to obtain mutant BW370, which has Bowman's genetic background but exhibits semi-dwarf and multi-tiller traits.
[0045] The nucleotide sequence of the wild-type Bowman HORVU2Hr1G098820 gene is described in SEQ ID NO: 2, and the protein encoded by it has the amino acid sequence described in SEQ ID NO: 4.
[0046] 2) Obtaining the molecular marker of the semi-dwarf and multi-tiller gene fol-a: According to the sequence comparison res...
Embodiment 2
[0052] Embodiment 2, identify the sequence difference of mutant BW370 and Bowman with molecular marker:
[0053] 1), DNA extraction: Take 100 mg of barley leaves, and use the CTAB method to extract DNA. The specific steps are as follows:
[0054] ① Grind the sheared leaves in liquid nitrogen and put them into 1.5ml centrifuge tubes, add 600 μL of CTAB to each tube and place in a warm water bath at 65°C for 50-60 minutes (take out every 10 minutes and shake gently);
[0055] ②Add 600 microliters of chloroform / isoamyl alcohol (24:1) and mix well for 15 minutes. Slowly, weigh in pairs, and after balancing, centrifuge at 9600 rpm for 10 minutes at 4°C;
[0056] ③Pipe the supernatant to another 1.5ml new centrifuge tube, add 2 times the volume of absolute ethanol and mix slightly (precipitate in -20°C refrigerator for 30-60min);
[0057] ③ Pick out the precipitate with a pipette tip, put it in a 1.5ml centrifuge tube, wash it twice with 70% ethanol and air dry;
[0058] ④ Add 100...
Embodiment 3
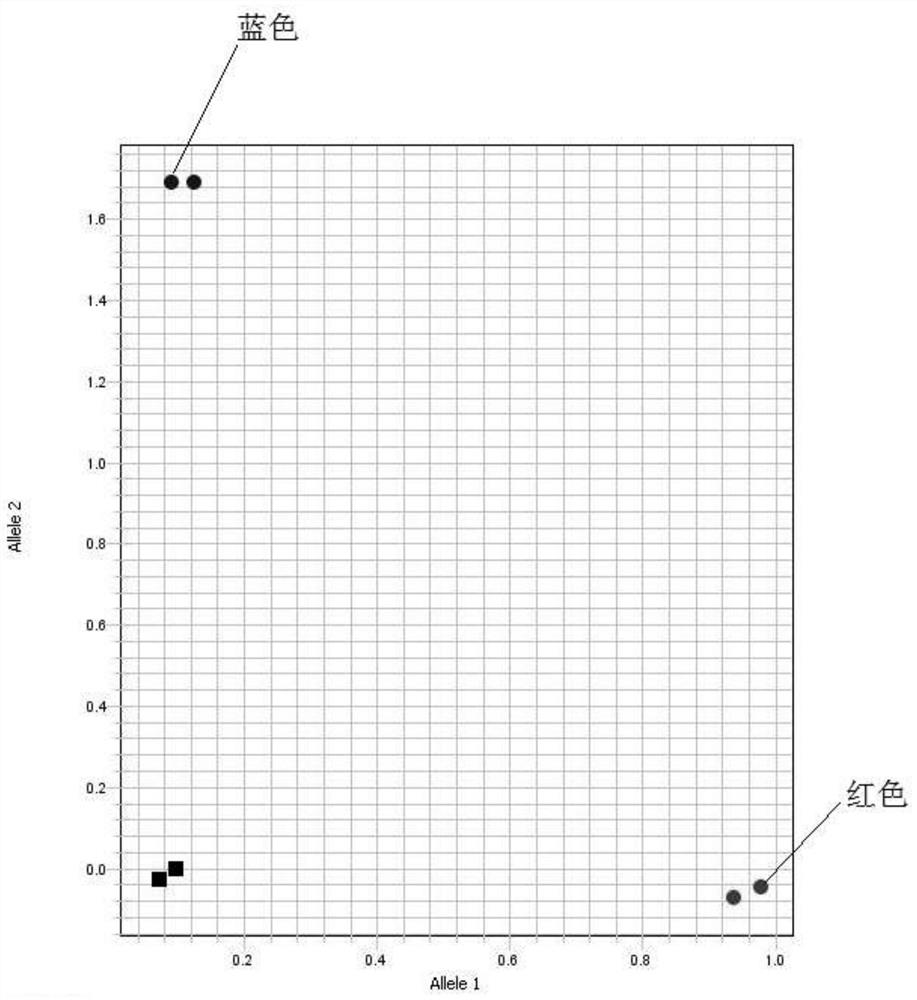
[0067] Example 3. Using the molecular marker SNPE3 to detect 32 barley germplasm resources (including mutants BW370 and Bowman, 2 repetitions) according to the method described in Example 2, the barley germplasm resources include wild barley, local varieties and from all over the world The barley cultivars and the F 1 (See Table 1);
[0068] The result is as Figure 4 shown.
[0069] The test results show that the F of BW370 / Vlamingh in these materials 1 The hybrid is heterozygous, and the mutant BW370 is the mutant genotype; other materials are all wild-type genotypes, and their phenotypes are also different from the semi-dwarf multi-tiller phenotype of the mutant BW370, indicating that they are not Containing the Hvthd mutant gene also proves that this molecular marker can be used in the detection of barley germplasm resources.
[0070] Table 1. Information on 32 barley germplasm resources detected by molecular marker SNPE3
[0071]
[0072]
[0073] Verification ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com