Molecular markers and methods for identifying Zhuxing three-piece fish and three-piece fish
A technique for molecular labeling and identification of beads, which is applied in biochemical equipment and methods, measurement/inspection of microorganisms, DNA/RNA fragments, etc., and can solve the problems of expensive capillary electrophoresis analysis process, cumbersome effective analysis and identification of alleles, and difficult implementation Problems such as complex process, to achieve the effect of protecting germplasm resources, fast identification process and strong operability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
specific Embodiment approach 1
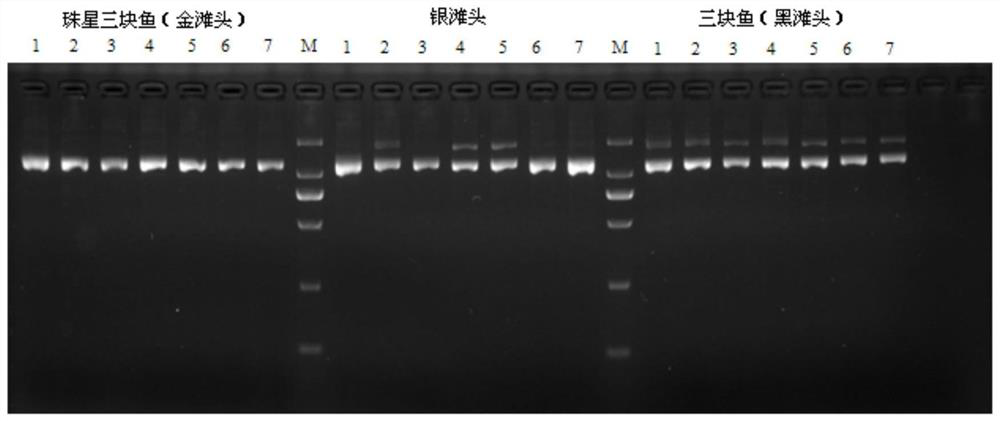
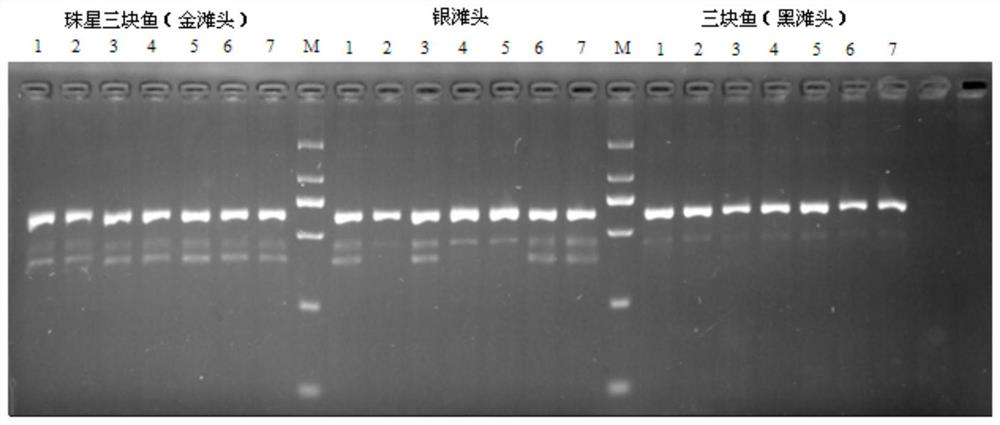
[0020] Specific embodiment 1: In this embodiment, the first molecular marker primer pair for identifying Zhuxing three-piece fish and three-piece fish is A8, and the nucleotide sequence of the A8 forward primer is 5'-GGCTACAGCAAGGCAAGAAC-3'; the A8 reverse primer The nucleotide sequence is 5'-ATGCATTTCCCTTCGTATGC-3'.
specific Embodiment approach 2
[0021] Specific embodiment 2: The second molecular marker primer pair for identifying Zhuxing three-piece fish and three-piece fish in this embodiment is A11, and the nucleotide sequence of the A11 forward primer is 5'-GGACTGGGGAGGTGTGAGTA-3'; the A11 reverse primer The nucleotide sequence is 5'-TGCAGAAAGAGCAGCAGAAA-3'.
specific Embodiment approach 3
[0022] Specific implementation mode three: the method for identification of Zhuxing three-piece fish and three-piece fish in this embodiment:
[0023] 1. Extraction of sample DNA;
[0024] 2. PCR detection:
[0025] The PCR reaction system is 25 μl, consisting of 18 μl self-made PCR buffer, 1.0 μl forward detection primer with a concentration of 10 μmol / L, 1.0 μl reverse detection primer with a concentration of 10 μmol / L, 2 μl sample DNA, and 0.2 μl Taq with an enzyme activity of 5 U DNA polymerase and the rest of deionized sterile water; 18 μl self-made PCR buffer containing 50mmol / L of KCl, 10mmol / L of Tris-HCl, 0.1% volume of TritonX-100, 1.5mmol / L of MgCl 2 , 0.1% by volume of NP-40, 0.01% by volume of gelatin and 200 μmol / L of four kinds of d NTP mixture; wherein the detection primers are molecular marker primer pair A8 or molecular marker primer pair A11;
[0026] The amplification conditions of PCR are: pre-denaturation at 95°C for 3min, denaturation at 95°C for 15s, ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com