Chitinase derived from marine environment and gene thereof
A chitinase gene, chitinase technology, applied in genetic engineering, plant genetic improvement, introduction of foreign genetic material using vectors, etc., can solve the problem of uncultivable microorganisms, etc. The effect of lasting stability
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0032] Embodiment 1 The acquisition of Chitinase gene
[0033] Operate according to the genome walking kit.
[0034] (1) Extraction of metagenomic DNA from mangrove tidal flat soil samples: OMEGA Soil DNA kit was used to extract metagenomic DNA from soil samples.
[0035] (2) Acquisition of chitinase conserved sequence
[0036] The Chitinase gene of subfamily A of the 18 families already available at NCBI was analyzed, and degenerate primers were designed to amplify the conserved sequence of the Chitinase gene. The degenerate primer sequences are as follows:
[0037] ChiA F: 5' GGTGGACATCGACTGGGARTWYCC 3';
[0038] ChiA R: 5' CCCAGGCGCCGTAGARRRTCRTARSWCA 3'.
[0039] The PCR program was: pre-denaturation at 95°C for 5 minutes; denaturation at 94°C for 30 s, annealing at 50°C for 30 s, extension at 72°C for 1 min, cycle amplification 35 times; final extension at 72°C for 10 min. After the amplification, the PCR product was taken for electrophoresis detection, and the target ...
Embodiment 2
[0055] Example 2 Expression and amplification of Chitinase coding gene in Escherichia coli
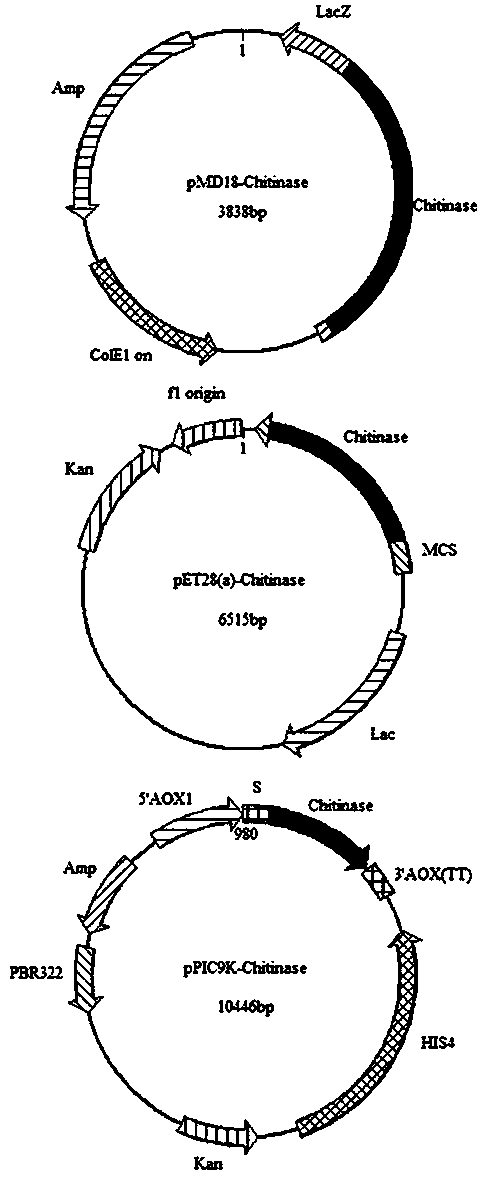
[0056] With the target gene obtained in embodiment 1-(5), and through to Sac I and xho The pET28a (+) plasmid of 1 double digestion is connected, obtains the recombinant plasmid pET28a (+)-Chitinase (such as figure 1 shown).
[0057] Take 10uL of the constructed plasmid DNA, add it to 100uL of the prepared competent Escherichia coli BL21(DE3), shake well and place it on ice for 30min; place it in a water bath at 42°C for 45s; place the centrifuge tube quickly Transfer to ice-water mixture for ice bath for 2min; add 800uL LB medium to each tube, lightly aspirate and disperse with a pipette, then recover on a shaker at 37°C for 1h (80rpm-200rpm); centrifuge, 4000rpm×5min, remove 700uL supernatant , and mix the remaining part; smear the plate (LB-agar plate, containing 100ug / mLAmp), place it upright at 37°C for 1 hour, and culture it upside down overnight. The positive clones contain...
Embodiment 3
[0059] Example 3 Purification of recombinant Chitinase
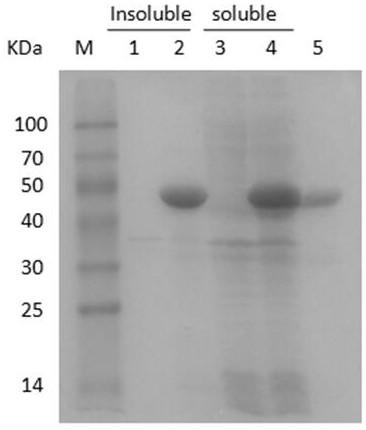
[0060] The fermentation broth prepared in Example 2 was centrifuged at 12,000 rpm for 10 min, and then the cells were resuspended in 20 mM Tris-HCl buffer (pH 7.4) and ultrasonically disrupted. The cells were broken and centrifuged at 12000rpm for 10min, and the supernatant was collected. Dilute the crude enzyme solution 5 times with buffer A (20mM Tris-HCl, 20mM imidazole, 500mM NaCl, pH 7.4), load it on a nickel column at a flow rate of 1mL / min, and use buffer A to elute weakly bound proteins to Equilibrate the column, then use buffer B (20mM Tris-HCl, 200mM imidazole, 500mM NaCl, pH 7.4) to elute the target protein at a flow rate of 1mL / min, collect the eluate, and measure the purity of the target protein by SDS-PAGE.
[0061] After the purification was completed, the specific activity of the obtained chitinase increased from 0.14 U / mg of the crude enzyme solution to 0.63 U / mg of the pure enzyme, the purification fac...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com