Polypeptides capable of forming homo-oligomers with modular hydrogen bond network-mediated specificity and their design
A technology of hydrogen bonding and network, applied in the field of computer readable media, computing equipment, protein oligomers, and devices, can solve the problems of polar amino acids that cannot interact with each other, such as hydrogen bonding capabilities
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
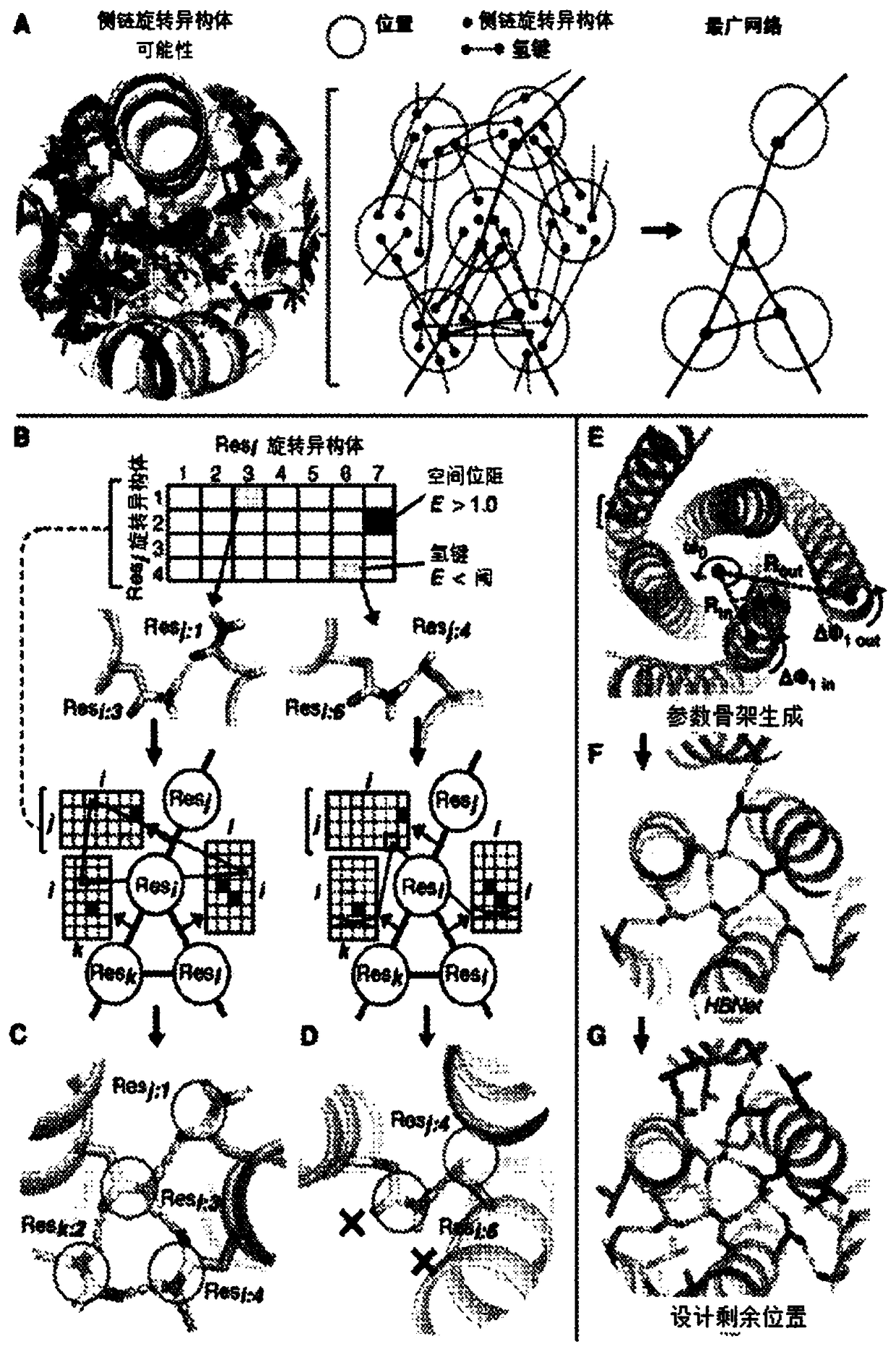
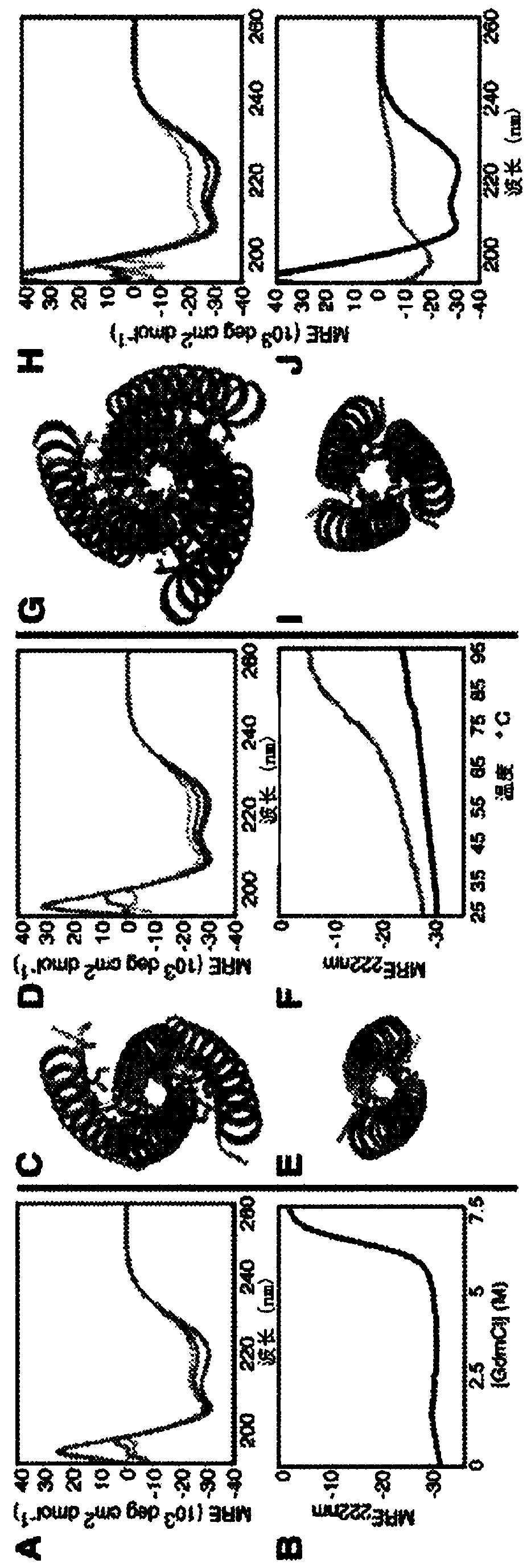
[0255] Modularity and predictability of DNA interaction specificity are at the heart of molecular biology manipulation and DNA nanotechnology, but there is no similarity in nature, and how to achieve similar programmable specificity with proteins is unclear. There are more polar amino acids than DNA bases, and each amino acid can adopt multiple side chain conformations in different backbone environments, enabling countless network possibilities. The inventors have developed a general computing method, the development of HBNet TM To quickly enumerate all possible side chain hydrogen bond networks in the input backbone structure (Figure IA).
[0256] Traditional protein design algorithms are not well suited for this purpose, since the total system energy usually represents the sum of interactions between pairs of residues used to calculate efficiencies, and thus cannot clearly disconnect a connected hydrogen bond network from a set of separate the hydrogen bonds. HBNet TM Fir...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com