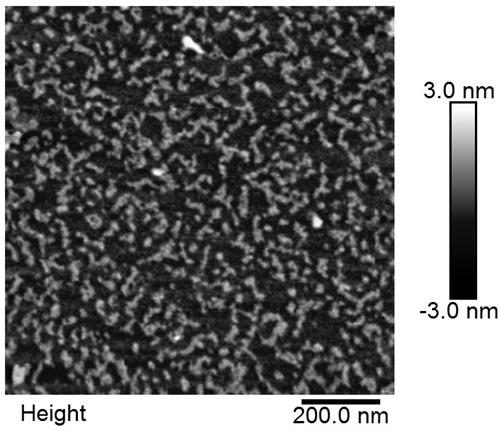
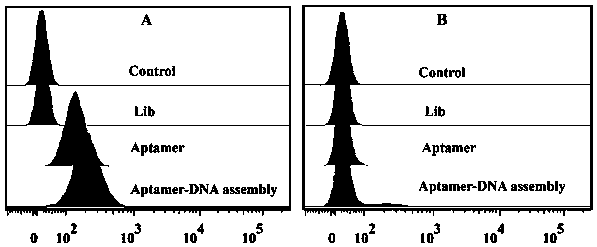
Construction method and application of aptamer-DNA macromolecular polymer based on non-linear hybridization chain amplification
A high molecular polymer, hybrid chain technology, applied in the construction and application of DNA high molecular polymers, can solve the problems of low bioavailability, strong side effects, non-targeting, etc., and achieve the effect of increasing application potential
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0043] Example 1 Synthesis of DNA polymers based on nonlinear HCR
[0044] (1) After dissolving the substrate strands S1 and S3, use a micro-ultraviolet spectrophotometer Q5000 to accurately measure the concentration, treat with 1uL phosphorylase, react in a PCR instrument at 37°C for 3h, and then treat at 95°C for 10min to end the phosphorylation process. Obtain base strands S1* and S3*;
[0045] (2) The bottom strands S1* and S2, and the bottom strands S3* and S4 were mixed at a molar ratio of 1:2, annealed in a PCR instrument at 90°C for 10 minutes, and cooled to 0.1°C / s to At 25°C, the main chain S1*S2 and the main chain S3*S4 were obtained, and stored at 4°C for later use;
[0046] (3) Auxiliary chains H1 and H2 are added to the solution of main chain S1*S2 and main chain S3*S4 respectively, the molar ratio of auxiliary chain H1 to auxiliary chain H2 is 1:2, and the ratio of auxiliary chain H1 to main chain S1*S2 The molar ratio is 1:3~2:3, and incubated in a PCR instru...
Embodiment 2
[0051] (1) After dissolving the base strands S1 and S3, use a micro-ultraviolet spectrophotometer Q5000 to accurately measure the concentration, heat in a PCR instrument at 37°C for 3 hours, and then treat at 95°C for 10 minutes to obtain the base strands S1* and S3*;
[0052] (2) The bottom strands S1* and S2, and the bottom strands S3* and S4 were mixed at a molar ratio of 1:2, annealed in a PCR instrument at 90°C for 10 minutes, and cooled to 0.1°C / s to At 25°C, the main chain S1*S2 and the main chain S3*S4 were obtained, and stored at 4°C for later use;
[0053] (3) Auxiliary chains H1 and H2 are added to the solution of main chain S1*S2 and main chain S3*S4 respectively, the molar ratio of auxiliary chain H1 to auxiliary chain H2 is 1:2, and the ratio of auxiliary chain H1 to main chain S1*S2 The molar ratio is 1:3~2:3, and incubated in a PCR instrument at 37°C for 30min;
[0054] (4) Mix the two solutions in step (3) evenly, add the initiator chain, the amount of the in...
Embodiment 3
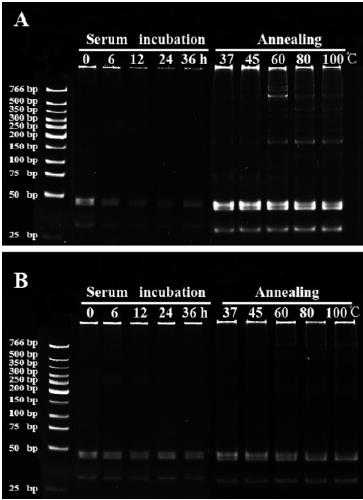
[0058] The gap-unclosed DNA polymer obtained in Example 1 and the gap-closed DNA polymer obtained in Example 2 were respectively reacted in a PCR instrument at 37°C for 0-36h, followed by polyacrylamide gelation. Gel electrophoresis detection, see the results figure 2 , figure 2 It is the measurement diagram of serum stability and temperature stability of two different DNA polymers (A: DNA polymer with open gap; B: DNA polymer with gap closed). It can be seen from the figure that the unlinked DNA polymer was degraded to a certain extent after 12 hours of serum incubation, and the degree of degradation gradually increased with the extension of incubation time, and most of the samples blocked in the gel pores had been degraded at 36 hours . In contrast, the linked DNA polymer has almost no effect, even if the incubation time reaches 36h, it still cannot degrade it.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com