Probability statistics model-based biological full-sibling identification method
A technology of probability and statistics models and identification methods, applied in the fields of bioinformatics, biological systems, informatics, etc., can solve problems such as the inability to study and evaluate probability distributions, and the lack of system effectiveness evaluation methods
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0141] According to the "Implementation Standards for Identification of Biological Full-Sibling Relationship, SF / Z JD0105002-2014", and adopt the biological full-sibling identification method based on the probability and statistics model described in the present invention to detect and identify the two identified persons:
[0142] Step 1, DNA extraction:
[0143] Use a commercially available DNA extraction kit to extract the finger blood genomic DNA of two identified persons;
[0144] Step 2, STR typing:
[0145] Use the Goldeneye 20A kit to carry out PCR amplification, and perform STR typing on the 19 must-test loci of the two identified persons to obtain the typing results of 19 mutually independent STR loci;
[0146] Step 3, establish the test hypothesis :
[0147] Null hypothesis H 0 : The two identified individuals are unrelated individuals;
[0148] Alternative Hypothesis H 1 : The two identified persons are biological compatriots;
[0149] Step 4, calculat...
Embodiment 2
[0161] Using the biological full sibling identification method based on the probability and statistics model described in the present invention to evaluate the effectiveness of the given threshold in the "Implementation Specification for Biological Full Sibling Relationship Identification, SF / Z JD0105002-2014":
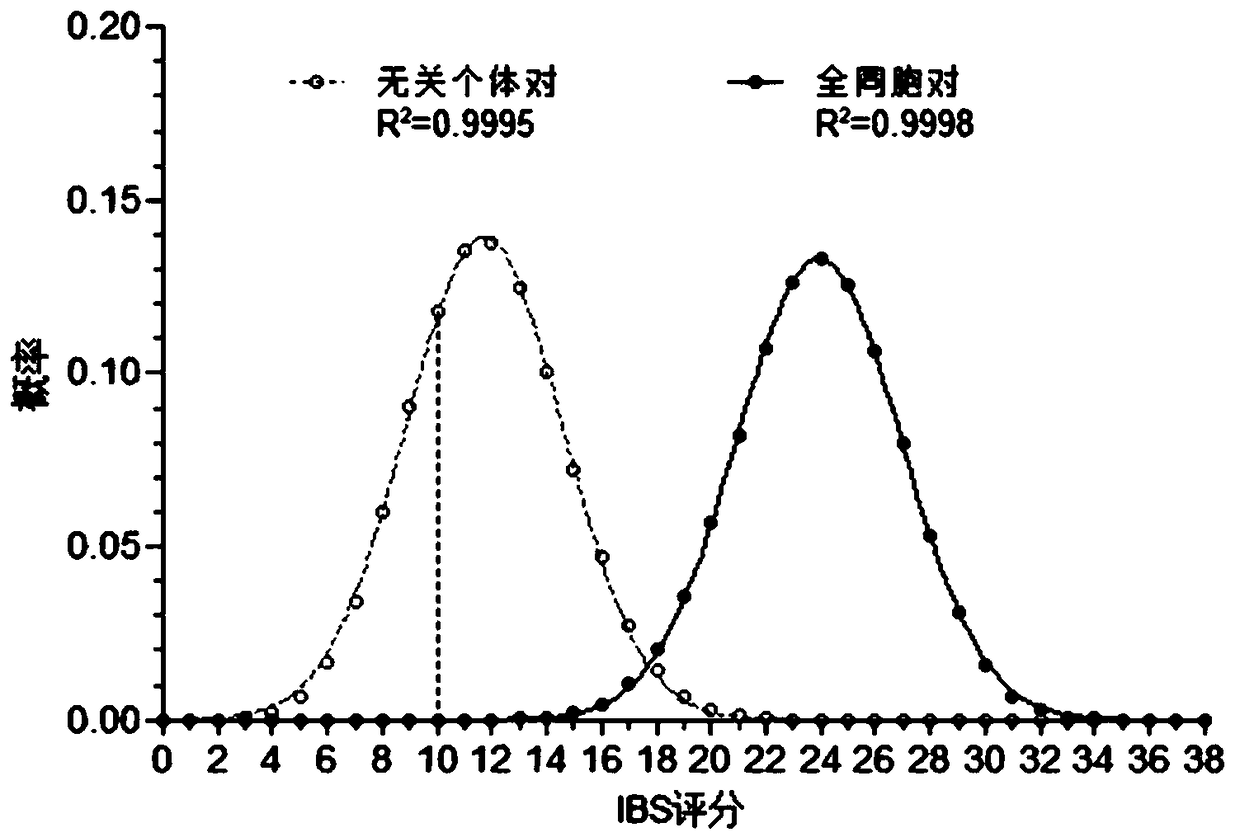
[0162] According to the above Example 1, when the 19 mandatory STR loci in the "Implementation Standards for the Identification of Biological Sibling Relationships, SF / ZJD0105002-2014" are used to perform the STR typing test on two identified persons, the IBS in the unrelated If the relational individual fits the binomial distribution IBS~B(38,0.3110) in the population, and the IBS fits the binomial distribution IBS~B(38,0.6280) in the whole sibling pair population, then:
[0163] The true unrelated individual with IBS<=13 is: BINOMDIST(13,38,0.3110,TURE)=0.7268
[0164] Full siblings with IBS>=22 are: 1-BINOMDIST(21,38,0.6280,TURE)=0.7876
[0165] Full siblings with...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com