CAPS molecular marker for identifying background color of watermelon peel and application thereof
A molecular marker and peel technology, used in recombinant DNA technology, microbial determination/inspection, DNA/RNA fragments, etc., can solve the problems of being easily affected by the environment and low efficiency, saving manpower and material resources, high sensitivity, and improving efficiency Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0029] Embodiment 1, the development of CAPS molecular markers and primers for identifying watermelon peel background color
[0030] Firstly, the fine mapping of skin color-related genes was carried out by constructing a high-density genetic map, as follows: by resequencing 126 individual plants of recombinant inbred lines and their parents (9904 as the female parent and Handel as the male parent), a A high-density genetic map of watermelon. A total of 178,762 SNP markers suitable for RILs with a depth of not less than 4X were detected between the parents. The total length of the map is 1508.94cM, including 11 linkage groups, 2132 Bin markers, and the average map distance is 0.74cM . Through bioinformatics analysis, the candidate genes that control the traits of peel background color were located in the range of 2.07Mb at the end of chromosome 8. By designing CAPS markers and SNP markers for fine mapping, the gene controlling the background color of watermelon peel was locate...
Embodiment 2
[0036] Embodiment 2, utilize above-mentioned CAPS molecular marker to identify the method for the background color of watermelon peel
[0037] 1. Extraction of genomic DNA from watermelon samples
[0038] Use conventional CTAB method to extract genomic DNA from watermelon leaves, remove RNA contamination, use UV spectrophotometer to detect OD values of DNA samples at 260nm and 280nm, and measure DNA concentration and OD 260 / 280 , choose OD 260 / 280 For high-quality DNA samples at 1.8-2.0, the concentration was diluted to 100ng / μL.
[0039] 2. PCR amplification
[0040] The above-mentioned watermelon sample genomic DNA was amplified with the primer set used to amplify the CAPS molecular marker in Example 1; the reaction system for PCR amplification was: 2×PCR Mix 12.5 μL, DNA 2 μL, upstream primer 1.25 μL, downstream Primer 1.25 μL, add deionized water to 25 μL.
[0041] The reaction program of PCR is: 94°C 1min30sec; 94°C 20sec, 57°C 20sec, 72°C 50sec, a total of 30 cycle...
Embodiment 3
[0044] Embodiment 3, the verification of the above-mentioned CAPS molecular markers to the phenotype of the pericarp background color of 105 watermelon germplasm resources
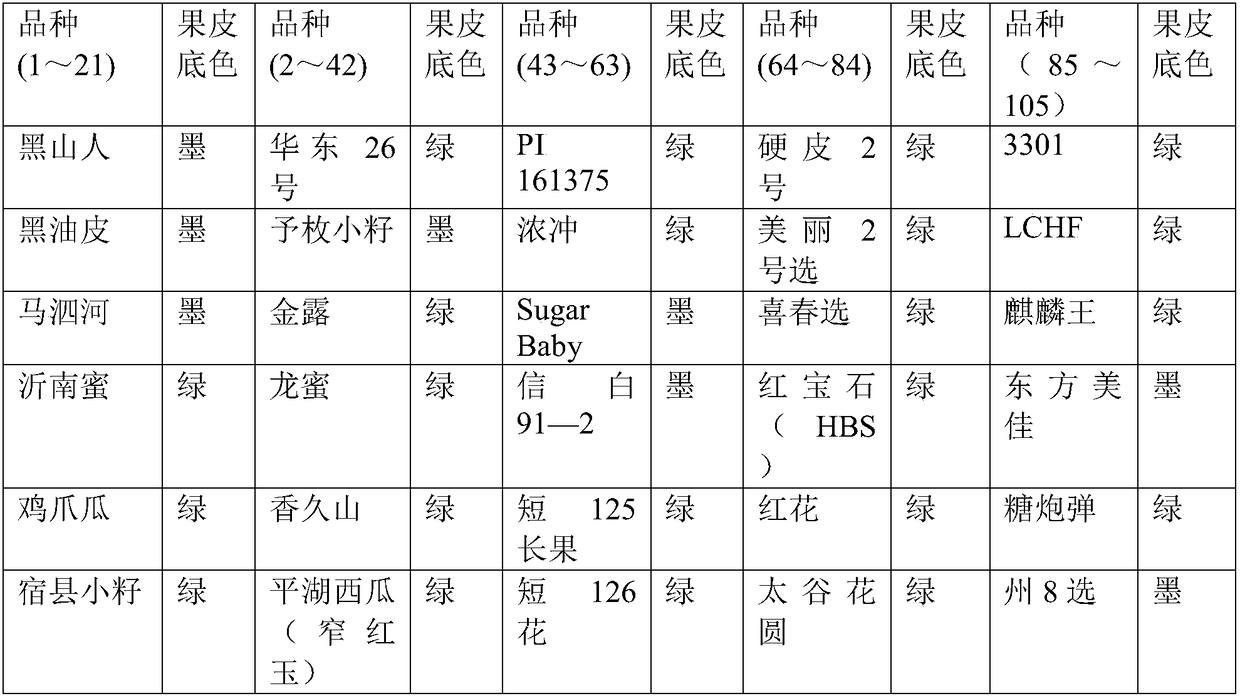
[0045] Taking 105 parts of watermelon germplasm resources collected by Zhengzhou Institute of Fruit Trees, Chinese Academy of Agricultural Sciences as material, using the CAPS molecular markers obtained by the present invention, adopting the method in Example 2 to carry out pericarp background color identification of 105 parts of watermelon germplasm resources, agar Glycogel electrophoresis results are shown in figure 1 . The identification results showed that the phenotypes of 105 watermelon germplasm resources were consistent with the band types of electrophoresis results, and the accuracy was 100%. Therefore, the CAPS molecular marker of the present invention can be used to identify the background color of watermelon peel.
[0046] Among the above-mentioned 105 germplasm resources, there are 31 waterm...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com