Application of keratinase gene of radiation-resistant gobi dysplasia coccus
A protease, gene technology, applied in the application, genetic engineering, plant genetic improvement and other directions, can solve the problem of not being able to play the role of enzyme activity, limiting the scope of use of alkaline protease, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1 Expression of Deinococcus Gobi KerA Gene Sequence in Escherichia coli
[0031] 1. Experimental materials
[0032] Escherichia coli BL21(DE3): It is a commercial product of Beijing Quanshijin Company.
[0033] PCR Template DNA: Deinococcus Gobi Genomic DNA
[0034] 2. Experimental method
[0035] 1. Design 1 pair of PCR-specific primers according to the KerA gene sequence in the published Deinococcus gobi genome:
[0036] KerA-F: 5′ACCGGATCCGATGAACGGACGTCTTACCCT 3′
[0037] KerA-R: 5′ACCCTCGAGGAAGTTCAGGGTGTACAGCA 3′
[0038] 2. The target gene sequence was amplified from the genomic DNA of Deinococcus gobii by PCR method.
[0039] Reaction conditions: 94°C for 10 min, 35 cycles of [94°C for 30 sec, 60°C for 30 sec, 72°C for 1.5 min], 72°C for 10 min.
[0040] 3. After the PCR product was recovered by gel, it was cloned on the vector pJET, named pJET-KerA, and verified by sequencing; then the KerA gene containing sticky ends and the pET-KerA gene containin...
Embodiment 2
[0045] Example 2 Preliminary Analysis of KerA Protease Activity of Deinococcus Gobi
[0046] 1. Experimental materials
[0047] Recombinant engineering strain: the BL21-KerA strain expressing the KerA gene obtained in Example 1
[0048] Control strain: the BL21-22b strain containing the empty plasmid described in Example 1.
[0049] 2. Experimental method
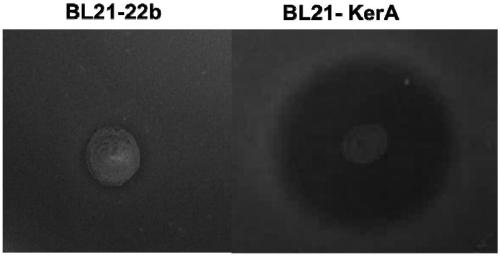
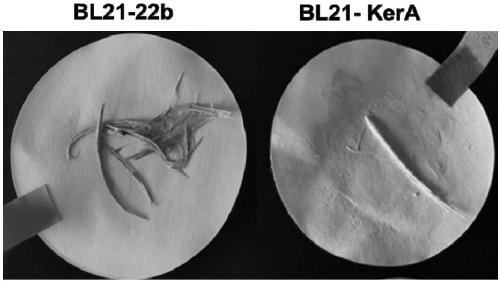
[0050] 1. Plate degradation circle experiment
[0051] 1) Pick out the monoclonal strains and inoculate them in liquid LB medium containing Amp, and culture them on a constant temperature shaker at 37°C overnight
[0052] 2) Transplant the seed liquid into a new LB medium, and cultivate to the logarithmic growth phase. Prepare 1% agar plates containing 1% skim milk. Spot the strain on the plate, measure the diameter of the hydrolysis circle and the diameter of the colony every 12 hours until it is stable.
[0053] 3) Calculate the ratio H / C of the diameter of the hydrolysis circle to the diameter of the colony on the ...
Embodiment 3
[0061] Example 3 Deinococcus Gobi KerA Protease Activity Assay Experiment
[0062] 1. Experimental materials
[0063] Recombinant engineering strain: the BL21-KerA strain expressing the KerA gene obtained in Example 1
[0064] 2. Experimental method
[0065] 1. Protein expression and purification
[0066] 1) Pick out the single clone and inoculate it into 50mL liquid LB medium containing Amp, and cultivate overnight in a constant temperature shaker at 37°C
[0067] 2) Transfer 0.1% of the seed solution of the transferred strain to fresh LB medium, and culture it on a constant temperature shaker at 37°C to OD 600 up to 0.6
[0068] 3) Add IPTG with a final concentration of 1.0mM to the medium, and induce culture at 30°C for 8h
[0069] 4) Centrifuge at 12000rpm to collect the bacteria and culture supernatant, the supernatant is the crude enzyme solution
[0070] 5) Add loading buffer, boil for SDS-PAGE analysis.
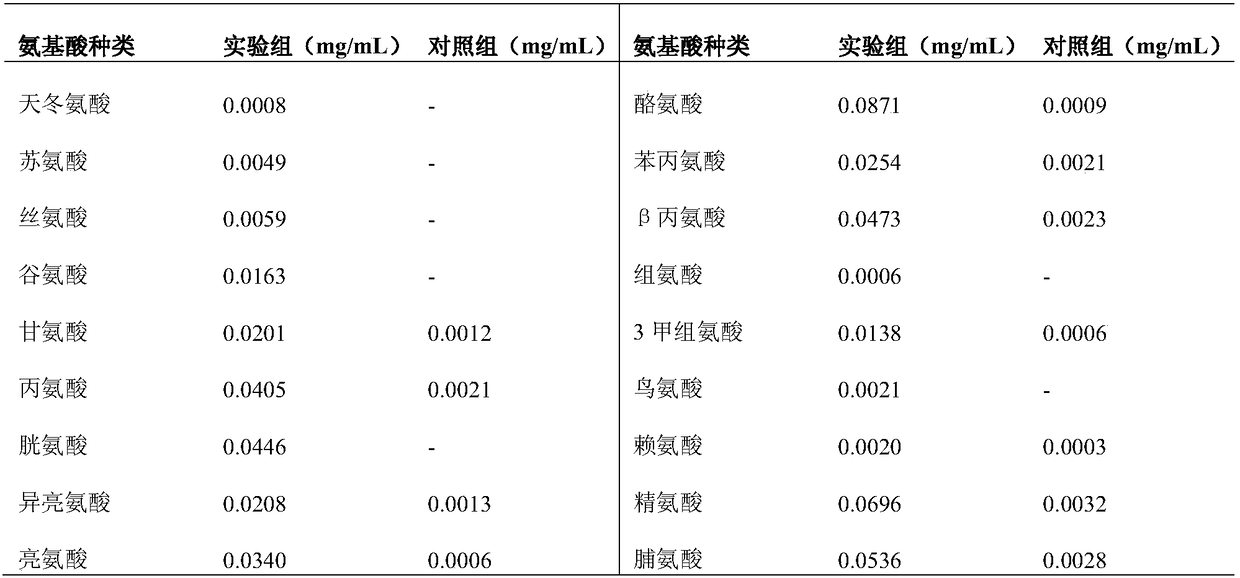
[0071] 2. Protease activity assay
[0072] The enzymatic...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com