DNA probe for enriching low-frequency DNA mutations and application of DNA probe
A DNA probe and low-frequency technology, applied in the direction of DNA/RNA fragments, recombinant DNA technology, microbial measurement/inspection, etc., can solve the problems of high sample size and limit the application of simultaneous detection of multiple sites, and achieve Efficient enrichment, significant enrichment, and dynamic monitoring
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
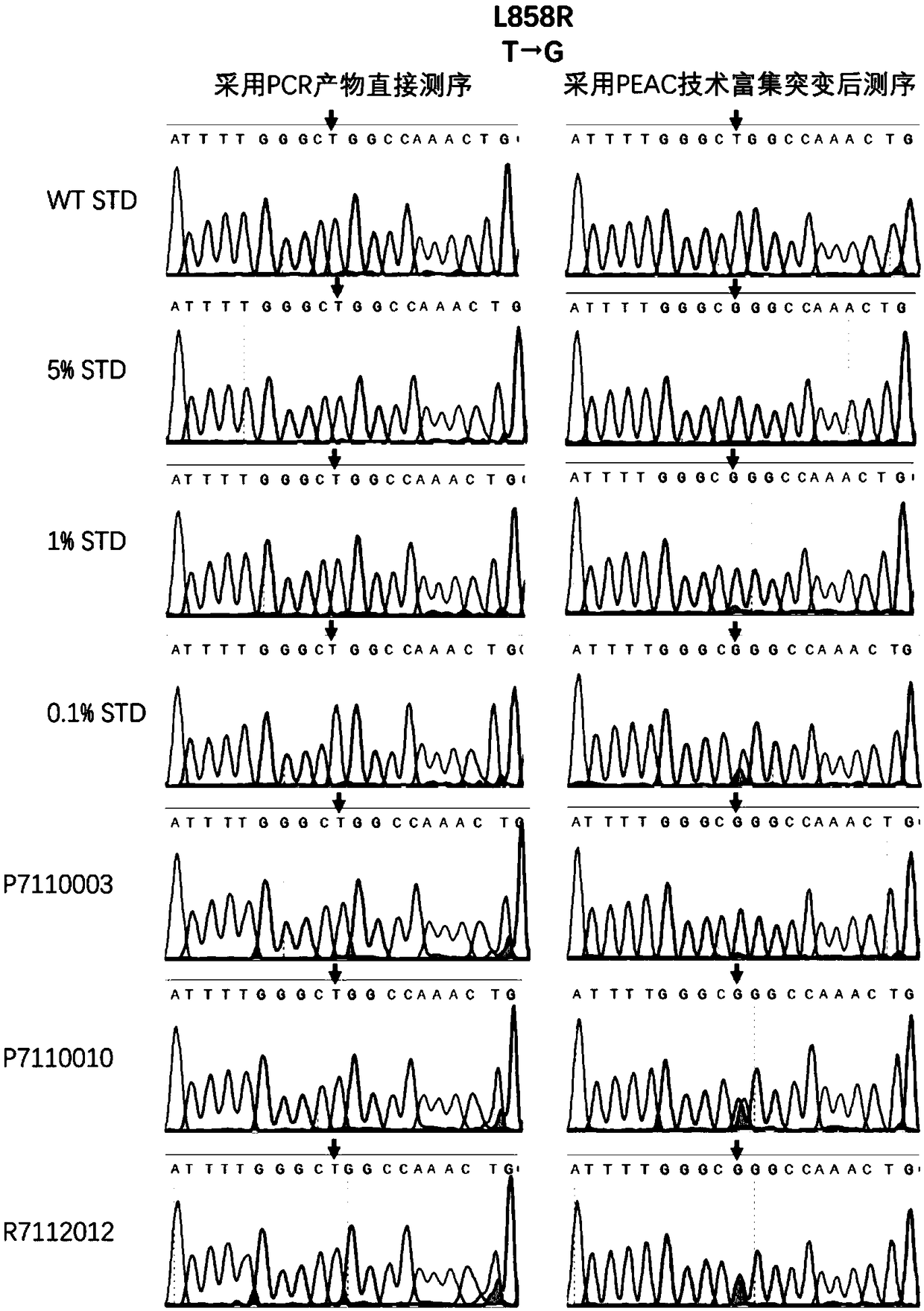
[0046] In this example, the L858R mutation on exon 21 of the EGFR gene hotspot mutation is taken as an example to analyze the method for capturing and enriching EGFR mutations by special DNA probes of the present invention. The samples used in the experiment were Horizon’s Multiplex I cfDNAReference Standard Set (HD780), which were EGFR gene wild type, with 5% EGFR L858R mutation, with 1% EGFR L858R mutation, and with 0.1% EGFR L858R mutation; 3 cases of lung adenocarcinoma Clinical samples.
[0047] The method for detecting the L858R mutation on exon 21 of the EGFR gene hotspot mutation by using the above method comprises the following steps:
[0048] (1) Separation of peripheral blood samples
[0049] 1. Collect one tube (10mL / tube) of the subject’s peripheral blood into a Streck tube, gently upside down (to prevent cell rupture) 6-8 times and mix well, and perform the following treatment within 7 days after blood collection;
[0050] 2. Centrifuge at 1600g for 10 minutes ...
Embodiment 2
[0109] In this example, the T790M mutation on exon 20 of the EGFR gene hotspot mutation is taken as an example to analyze the method for capturing and enriching EGFR mutations by special DNA probes of the present invention. The samples used in the experiment were Horizon’s Multiplex I cfDNAReference Standard Set (HD780), which were EGFR gene wild type, with 5% EGFR T790M mutation, with 1% EGFR T790M mutation, and with 0.1% EGFR T790M mutation; two cases of lung adenocarcinoma Clinical samples.
[0110] The method for detecting the T790M mutation on exon 20 of the EGFR gene hotspot mutation by using the above method comprises the following steps:
[0111] (1) Separation of peripheral blood samples
[0112] 1. Collect 1 tube (10mL / tube) of peripheral blood from the subject into a Streck tube, gently invert it upside down (to prevent cell rupture) 6-8 times and mix thoroughly, and perform the following treatment within 7 days after blood collection;
[0113] 2. Centrifuge at 16...
Embodiment 3
[0173] In this example, the deletion mutation on exon 19 of the EGFR gene hotspot mutation is taken as an example to analyze the method for capturing and enriching EGFR mutation with a special DNA probe of the present invention. The samples used in the experiment were Horizon’s Multiplex I cfDNAReference Standard Set (HD780), which were EGFR gene wild type, 5% EGFR 19DEL mutation, 1% EGFR 19DEL mutation, and 0.1% EGFR 19DEL mutation; two cases of lung adenocarcinoma Clinical samples.
[0174] The method for detecting the deletion mutation on exon 20 of EGFR gene hotspot mutation by using the above method comprises the following steps:
[0175] (1) Separation of peripheral blood samples
[0176] 1. Collect 1 tube (10mL / tube) of peripheral blood from the subject into a Streck tube, gently invert it upside down (to prevent cell rupture) 6-8 times and mix thoroughly, and perform the following treatment within 7 days after blood collection;
[0177] 2. Centrifuge at 1600g for 10 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - Generate Ideas
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com